+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | INF2 in the middle of F-Actin, Focus refinement for mobile WH2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Actin / Formin / Filament / Severing / CYTOSOLIC PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Palmer NJ / Barrie KR / Dominguez R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Mechanisms of actin filament severing and elongation by formins. Authors: Nicholas J Palmer / Kyle R Barrie / Roberto Dominguez /  Abstract: Humans express 15 formins that play crucial roles in actin-based processes, including cytokinesis, cell motility and mechanotransduction. However, the lack of structures bound to the actin filament ...Humans express 15 formins that play crucial roles in actin-based processes, including cytokinesis, cell motility and mechanotransduction. However, the lack of structures bound to the actin filament (F-actin) has been a major impediment to understanding formin function. Whereas formins are known for their ability to nucleate and elongate F-actin, some formins can additionally depolymerize, sever or bundle F-actin. Two mammalian formins, inverted formin 2 (INF2) and diaphanous 1 (DIA1, encoded by DIAPH1), exemplify this diversity. INF2 shows potent severing activity but elongates weakly whereas DIA1 has potent elongation activity but does not sever. Using cryo-electron microscopy (cryo-EM) we show five structural states of INF2 and two of DIA1 bound to the middle and barbed end of F-actin. INF2 and DIA1 bind differently to these sites, consistent with their distinct activities. The formin-homology 2 and Wiskott-Aldrich syndrome protein-homology 2 (FH2 and WH2, respectively) domains of INF2 are positioned to sever F-actin, whereas DIA1 appears unsuited for severing. These structures also show how profilin-actin is delivered to the fast-growing barbed end, and how this is followed by a transition of the incoming monomer into the F-actin conformation and the release of profilin. Combined, the seven structures presented here provide step-by-step visualization of the mechanisms of F-actin severing and elongation by formins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44943.map.gz emd_44943.map.gz | 46 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44943-v30.xml emd-44943-v30.xml emd-44943.xml emd-44943.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_44943_fsc.xml emd_44943_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_44943.png emd_44943.png | 35.7 KB | ||
| Masks |  emd_44943_msk_1.map emd_44943_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-44943.cif.gz emd-44943.cif.gz | 4.3 KB | ||
| Others |  emd_44943_half_map_1.map.gz emd_44943_half_map_1.map.gz emd_44943_half_map_2.map.gz emd_44943_half_map_2.map.gz | 84.5 MB 84.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44943 http://ftp.pdbj.org/pub/emdb/structures/EMD-44943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44943 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44943.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44943.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_44943_msk_1.map emd_44943_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
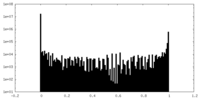
| Density Histograms |
-Half map: #1
| File | emd_44943_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
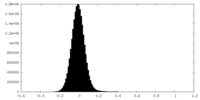
| Density Histograms |
-Half map: #2
| File | emd_44943_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
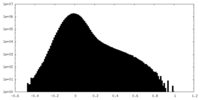
| Density Histograms |
- Sample components
Sample components
-Entire : INF2 in the Middle of Actin Filament
| Entire | Name: INF2 in the Middle of Actin Filament |
|---|---|
| Components |
|
-Supramolecule #1: INF2 in the Middle of Actin Filament
| Supramolecule | Name: INF2 in the Middle of Actin Filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Supramolecule #2: INF2
| Supramolecule | Name: INF2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Actin Filament
| Supramolecule | Name: Actin Filament / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.45 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 100 | ||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 41926 / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)