[English] 日本語
 Yorodumi
Yorodumi- EMDB-4460: Cryo-EM structure of Salmonella AcrB solubilised in the SMA copolymer -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4460 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Salmonella AcrB solubilised in the SMA copolymer | |||||||||
 Map data Map data | Local resolution filtered map of Salmonella AcrB | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationxenobiotic transport / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Salmonella (bacteria) Salmonella (bacteria) | |||||||||
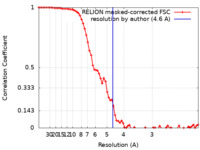
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Johnson RM / Bavro VN / Postis V / Muench SP | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Microorganisms / Year: 2020 Journal: Microorganisms / Year: 2020Title: Cryo-EM Structure and Molecular Dynamics Analysis of the Fluoroquinolone Resistant Mutant of the AcrB Transporter from . Authors: Rachel M Johnson / Chiara Fais / Mayuriben Parmar / Harish Cheruvara / Robert L Marshall / Sophie J Hesketh / Matthew C Feasey / Paolo Ruggerone / Attilio V Vargiu / Vincent L G Postis / ...Authors: Rachel M Johnson / Chiara Fais / Mayuriben Parmar / Harish Cheruvara / Robert L Marshall / Sophie J Hesketh / Matthew C Feasey / Paolo Ruggerone / Attilio V Vargiu / Vincent L G Postis / Stephen P Muench / Vassiliy N Bavro /   Abstract: is an important genus of Gram-negative pathogens, treatment of which has become problematic due to increases in antimicrobial resistance. This is partly attributable to the overexpression of ... is an important genus of Gram-negative pathogens, treatment of which has become problematic due to increases in antimicrobial resistance. This is partly attributable to the overexpression of tripartite efflux pumps, particularly the constitutively expressed AcrAB-TolC. Despite its clinical importance, the structure of the AcrB transporter remained unknown to-date, with much of our structural understanding coming from the orthologue. Here, by taking advantage of the styrene maleic acid (SMA) technology to isolate membrane proteins with closely associated lipids, we report the very first experimental structure of AcrB transporter. Furthermore, this novel structure provides additional insight into mechanisms of drug efflux as it bears the mutation (G288D), originating from a clinical isolate of Typhimurium presenting an increased resistance to fluoroquinolones. Experimental data are complemented by state-of-the-art molecular dynamics (MD) simulations on both the wild type and G288D variant of AcrB. Together, these reveal several important differences with respect to the protein, providing insights into the role of the G288D mutation in increasing drug efflux and extending our understanding of the mechanisms underlying antibiotic resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4460.map.gz emd_4460.map.gz | 20.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4460-v30.xml emd-4460-v30.xml emd-4460.xml emd-4460.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4460_fsc.xml emd_4460_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4460.png emd_4460.png | 156.6 KB | ||
| Others |  emd_4460_additional_1.map.gz emd_4460_additional_1.map.gz emd_4460_additional_2.map.gz emd_4460_additional_2.map.gz emd_4460_half_map_1.map.gz emd_4460_half_map_1.map.gz emd_4460_half_map_2.map.gz emd_4460_half_map_2.map.gz | 6.8 MB 23.2 MB 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4460 http://ftp.pdbj.org/pub/emdb/structures/EMD-4460 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4460 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4460 | HTTPS FTP |
-Validation report
| Summary document |  emd_4460_validation.pdf.gz emd_4460_validation.pdf.gz | 361 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4460_full_validation.pdf.gz emd_4460_full_validation.pdf.gz | 360.1 KB | Display | |
| Data in XML |  emd_4460_validation.xml.gz emd_4460_validation.xml.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4460 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4460 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4460 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4460 | HTTPS FTP |
-Related structure data
| Related structure data |  6z12MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4460.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4460.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
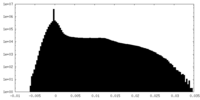
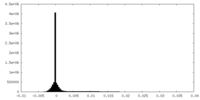
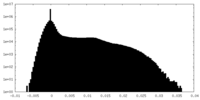
| Annotation | Local resolution filtered map of Salmonella AcrB | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Post-processed (sharpened) cryo-EM map of Salmonella AcrB
| File | emd_4460_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed (sharpened) cryo-EM map of Salmonella AcrB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened cryo-EM map of Salmonella AcrB
| File | emd_4460_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-EM map of Salmonella AcrB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 for Salmonella AcrB
| File | emd_4460_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 for Salmonella AcrB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 for Salmonella AcrB
| File | emd_4460_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 for Salmonella AcrB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Salmonella AcrB
| Entire | Name: Salmonella AcrB |
|---|---|
| Components |
|
-Supramolecule #1: Salmonella AcrB
| Supramolecule | Name: Salmonella AcrB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Cryo-EM structure of Salmonella multidrug bacterial efflux pump AcrB solubilised in SMA co-polymer |
|---|---|
| Source (natural) | Organism:  Salmonella (bacteria) Salmonella (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 340 KDa |
-Macromolecule #1: Salmonella multidrug efflux transporter AcrB in a SMALP platform
| Macromolecule | Name: Salmonella multidrug efflux transporter AcrB in a SMALP platform type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella (bacteria) Salmonella (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MPNFFIDRPI FAWVIAIIIM LAGGLAILKL PVAQYPTIAP PAVTISATYP GADAKTVQDT VTQVIEQNM NGIDNLMYMS SNSDSTGTVQ ITLTFESGTD ADIAQVQVQN KLQLAMPLLP Q EVQQQGVS VEKSSSSFLM VVGVINTDGT MTQEDISDYV AANMKDPISR ...String: MPNFFIDRPI FAWVIAIIIM LAGGLAILKL PVAQYPTIAP PAVTISATYP GADAKTVQDT VTQVIEQNM NGIDNLMYMS SNSDSTGTVQ ITLTFESGTD ADIAQVQVQN KLQLAMPLLP Q EVQQQGVS VEKSSSSFLM VVGVINTDGT MTQEDISDYV AANMKDPISR TSGVGDVQLF GS QYAMRIW MNPTELTKYQ LTPVDVINAI KAQNAQVAAG QLGGTPPVKG QQLNASIIAQ TRL TSTDEF GKILLKVNQD GSQVRLRDVA KIELGGENYD VIAKFNGQPA SGLGIKLATG ANAL DTATA IRAELKKMEP FFPPGMKIVY PYDTTPFVKI SIHEVVKTLV EAIILVFLVM YLFLQ NFRA TLIPTIAVPV VLLGTFAVLA AFGFSINTLT MFGMVLAIGL LVDDAIVVVE NVERVM TEE GLPPKEATRK SMGQIQGALV GIAMVLSAVF IPMAFFGGST GAIYRQFSIT IVSAMAL SV LVALILTPAL CATMLKPVAK GDHGEGKKGF FGWFNRLFDK STHHYTDSVG NILRSTGR Y LLLYLIIVVG MAYLFVRLPS SFLPDEDQGV FLTMVQLPAG ATQERTQKVL DEVTDYYLN KEKANVESVF AVNGFGFAGR GQNTGIAFVS LKDWADRPGE KNKVEAITQR ATAAFSQIKD AMVFAFNLP AIVELGTATG FDFELIDQAG LGHEKLTQAR NQLFGEVAKY PDLLVGVRPN G LEDTPQFK IDIDQEKAQA LGVSISDINT TLGAAWGGSY VNDFIDRGRV KKVYVMSEAK YR MLPDDIN DWYVRGSDGQ MVPFSAFSSS RWEYGSPRLE RYNGLPSMEI LGQAAPGKST GEA MAMMEE LASKLPSGIG YDWTGMSYQE RLSGNQAPAL YAISLIVVFL CLAALYESWS IPFS VMLVV PLGVIGALLA ATFRGLTNDV YFQVGLLTTI GLSAKNAILI VEFAKDLMDK EGKGL VEAT LEAVRMRLRP ILMTSLAFML GVMPLVISSG AGSGAQNAVG TGVLGGMVTA TVLAIF FVP VFFVVVRRRF SRKSEDIEHS HSTEHR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Salmonella AcrB was prepared in buffer containing 50mM Tris (pH8) with 500 mM Nacl and 10% glycerol. For EM grids, the sample was diluted into buffer containing 50 mM Tris and 150 mM NaCl. | |||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Grids were prepared using a Vitrobot Mark IV and blotted with Ash-free Whatman filter paper (No. 50) for 6 seconds using a blot force of 6.. | |||||||||
| Details | Salmonella AcrB was prepared in buffer containing 50mM Tris (pH8) with 500 mM Nacl and 10% glycerol. For EM grids, the sample was diluted into buffer containing 50 mM Tris and 150 mM NaCl. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 3210 / Average exposure time: 10.0 sec. / Average electron dose: 61.7 e/Å2 / Details: 3,210 images were collected in a 72 hour period. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.0045 µm / Nominal defocus min: 0.0015 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)