+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | CryoEM structure of DIM2-HP1 complex | |||||||||
 マップデータ マップデータ | sharpened map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | DNA methyltransferase / TRANSFERASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報regulation of biosynthetic process / DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase activity / negative regulation of gene expression via chromosomal CpG island methylation / heterochromatin / methylation / chromatin remodeling / chromatin binding / DNA binding / nucleus 類似検索 - 分子機能 | |||||||||
| 生物種 |  Neurospora crassa (菌類) Neurospora crassa (菌類) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.76 Å | |||||||||
 データ登録者 データ登録者 | Song J / Shao Z | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2024 ジャーナル: Nat Commun / 年: 2024タイトル: Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation. 著者: Zengyu Shao / Jiuwei Lu / Nelli Khudaverdyan / Jikui Song /  要旨: Functional crosstalk between DNA methylation, histone H3 lysine-9 trimethylation (H3K9me3) and heterochromatin protein 1 (HP1) is essential for proper heterochromatin assembly and genome stability. ...Functional crosstalk between DNA methylation, histone H3 lysine-9 trimethylation (H3K9me3) and heterochromatin protein 1 (HP1) is essential for proper heterochromatin assembly and genome stability. However, how repressive chromatin cues guide DNA methyltransferases for region-specific DNA methylation remains largely unknown. Here, we report structure-function characterizations of DNA methyltransferase Defective-In-Methylation-2 (DIM2) in Neurospora. The DNA methylation activity of DIM2 requires the presence of both H3K9me3 and HP1. Our structural study reveals a bipartite DIM2-HP1 interaction, leading to a disorder-to-order transition of the DIM2 target-recognition domain that is essential for substrate binding. Furthermore, the structure of DIM2-HP1-H3K9me3-DNA complex reveals a substrate-binding mechanism distinct from that for its mammalian orthologue DNMT1. In addition, the dual recognition of H3K9me3 peptide by the DIM2 RFTS and BAH1 domains allosterically impacts the DIM2-substrate binding, thereby controlling DIM2-mediated DNA methylation. Together, this study uncovers how multiple heterochromatin factors coordinately orchestrate an activity-switching mechanism for region-specific DNA methylation. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_44415.map.gz emd_44415.map.gz | 117.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-44415-v30.xml emd-44415-v30.xml emd-44415.xml emd-44415.xml | 18.3 KB 18.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_44415.png emd_44415.png | 67 KB | ||
| Filedesc metadata |  emd-44415.cif.gz emd-44415.cif.gz | 6.9 KB | ||
| その他 |  emd_44415_additional_1.map.gz emd_44415_additional_1.map.gz emd_44415_half_map_1.map.gz emd_44415_half_map_1.map.gz emd_44415_half_map_2.map.gz emd_44415_half_map_2.map.gz | 63.1 MB 116.1 MB 116.1 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44415 http://ftp.pdbj.org/pub/emdb/structures/EMD-44415 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44415 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44415 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_44415_validation.pdf.gz emd_44415_validation.pdf.gz | 748.1 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_44415_full_validation.pdf.gz emd_44415_full_validation.pdf.gz | 747.6 KB | 表示 | |
| XML形式データ |  emd_44415_validation.xml.gz emd_44415_validation.xml.gz | 13.8 KB | 表示 | |
| CIF形式データ |  emd_44415_validation.cif.gz emd_44415_validation.cif.gz | 16.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44415 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44415 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44415 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44415 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_44415.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_44415.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | sharpened map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.0515 Å | ||||||||||||||||||||||||||||||||||||
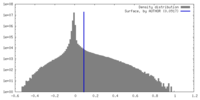
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: unsharpened map
| ファイル | emd_44415_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | unsharpened map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
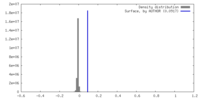
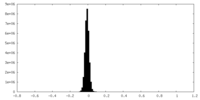
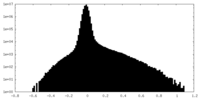
| 密度ヒストグラム |
-ハーフマップ: half map A
| ファイル | emd_44415_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | half map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
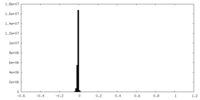
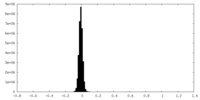
| 密度ヒストグラム |
-ハーフマップ: half map B
| ファイル | emd_44415_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | half map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
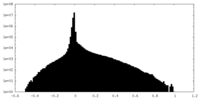
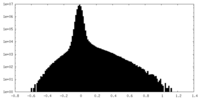
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : DIM2-HP1 complex
| 全体 | 名称: DIM2-HP1 complex |
|---|---|
| 要素 |
|
-超分子 #1: DIM2-HP1 complex
| 超分子 | 名称: DIM2-HP1 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#2 |
|---|---|
| 由来(天然) | 生物種:  Neurospora crassa (菌類) Neurospora crassa (菌類) |
-分子 #1: DNA (cytosine-5-)-methyltransferase
| 分子 | 名称: DNA (cytosine-5-)-methyltransferase / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO / EC番号: DNA (cytosine-5-)-methyltransferase |
|---|---|
| 由来(天然) | 生物種:  Neurospora crassa (菌類) Neurospora crassa (菌類) |
| 分子量 | 理論値: 139.935891 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSMDSPDRSH GGMFIDVPAE TMGFQEDYLD MFASVLSQGL AKEGDYAHHQ PLPAGKEECL EPIAVATTIT PSPDDPQLQL QLELEQQFQ TESGLNGVDP APAPESEDEA DLPDGFSDES PDDDFVVQRS KHITVDLPVS TLINPRSTFQ RIDENDNLVP P PQSTPERV ...文字列: GSMDSPDRSH GGMFIDVPAE TMGFQEDYLD MFASVLSQGL AKEGDYAHHQ PLPAGKEECL EPIAVATTIT PSPDDPQLQL QLELEQQFQ TESGLNGVDP APAPESEDEA DLPDGFSDES PDDDFVVQRS KHITVDLPVS TLINPRSTFQ RIDENDNLVP P PQSTPERV AVEDLLKAAK AAGKNKEDYI EFELHDFNFY VNYAYHPQEM RPIQLVATKV LHDKYYFDGV LKYGNTKHYV TG MQVLELP VGNYGASLHS VKGQIWVRSK HNAKKEIYYL LKKPAFEYQR YYQPFLWIAD LGKHVVDYCT RMVERKREVT LGC FKSDFI QWASKAHGKS KAFQNWRAQH PSDDFRTSVA ANIGYIWKEI NGVAGAKRAA GDQLFRELMI VKPGQYFRQE VPPG PVVTE GDRTVAATIV TPYIKECFGH MILGKVLRLA GEDAEKEKEV KLAKRLKIEN KNATKADTKD DMKNDTATES LPTPL RSLP VQVLEATPIE SDIVSIVSSD LPPSENNPPP LTNGSVKPKA KANPKPKPST QPLHAAHVKY LSQELVNKIK VGDVIS TPR DDSSNTDTKW KPTDTDDHRW FGLVQRVHTA KTKSSGRGLN SKSFDVIWFY RPEDTPCCAM KYKWRNELFL SNHCTCQ EG HHARVKGNEV LAVHPVDWFG TPESNKGEFF VRQLYESEQR RWITLQKDHL TCYHNQPPKP PTAPYKPGDT VLATLSPS D KFSDPYEVVE YFTQGEKETA FVRLRKLLRR RKVDRQDAPA NELVYTEDLV DVRAERIVGK CIMRCFRPDE RVPSPYDRG GTGNMFFITH RQDHGRCVPL DTLPPTLRQG FNPLGNLGKP KLRGMDLYCG GGNFGRGLEE GGVVEMRWAN DIWDKAIHTY MANTPDPNK TNPFLGSVDD LLRLALEGKF SDNVPRPGEV DFIAAGSPCP GFSLLTQDKK VLNQVKNQSL VASFASFVDF Y RPKYGVLE NVSGIVQTFV NRKQDVLSQL FCALVGMGYQ AQLILGDAWA HGAPQSRERV FLYFAAPGLP LPDPPLPSHS HY RVKNRNI GFLCNGESYV QRSFIPTAFK FVSAGEGTAD LPKIGDGKPD ACVRFPDHRL ASGITPYIRA QYACIPTHPY GMN FIKAWN NGNGVMSKSD RDLFPSEGKT RTSDASVGWK RLNPKTLFPT VTTTSNPSDA RMGPGLHWDE DRPYTVQEMR RAQG YLDEE VLVGRTTDQW KLVGNSVSRH MALAIGLKFR EAWLGTLYD UniProtKB: DNA (cytosine-5-)-methyltransferase |
-分子 #2: Heterochromatin protein one
| 分子 | 名称: Heterochromatin protein one / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Neurospora crassa (菌類) Neurospora crassa (菌類) |
| 分子量 | 理論値: 30.489086 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSMPYDPSAL SDEEAASSVE LDTRSATSSS KKQSRDKKSV KYTIPEPEDF EDEEQNGDGA DEGGEDDEEG DEEEEDVYVV EKILDHMLN DDNEPLFLVK WEGYEKKSDQ TWEPEDTLIE GASERLKEYF TKIGGREKIF EASAAAQKIK KRGRPSSNSG T PQASSNKR ...文字列: GSMPYDPSAL SDEEAASSVE LDTRSATSSS KKQSRDKKSV KYTIPEPEDF EDEEQNGDGA DEGGEDDEEG DEEEEDVYVV EKILDHMLN DDNEPLFLVK WEGYEKKSDQ TWEPEDTLIE GASERLKEYF TKIGGREKIF EASAAAQKIK KRGRPSSNSG T PQASSNKR SRKNGDHPLN SEEPQTAKNA AWKPPAGSWE EHIAQLDACE DEDTHKLMVY LTWKNGHKTQ HTTDVIYKRC PQ KMLQFYE RHVRIIKRDP DSEDREGSVS Q UniProtKB: Heterochromatin protein one |
-分子 #3: S-ADENOSYL-L-HOMOCYSTEINE
| 分子 | 名称: S-ADENOSYL-L-HOMOCYSTEINE / タイプ: ligand / ID: 3 / コピー数: 1 / 式: SAH |
|---|---|
| 由来(天然) | 生物種:  Neurospora crassa (菌類) Neurospora crassa (菌類) |
| 分子量 | 理論値: 384.411 Da |
| Chemical component information |  ChemComp-SAH: |
-分子 #4: ZINC ION
| 分子 | 名称: ZINC ION / タイプ: ligand / ID: 4 / コピー数: 1 / 式: ZN |
|---|---|
| 分子量 | 理論値: 65.409 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 0.8 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: NONE |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 2.76 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 296077 |
| 初期 角度割当 | タイプ: ANGULAR RECONSTITUTION |
| 最終 角度割当 | タイプ: ANGULAR RECONSTITUTION |
 ムービー
ムービー コントローラー
コントローラー










 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)