[English] 日本語
 Yorodumi
Yorodumi- EMDB-44369: Cryo-EM structure of the human TRPM4 channel in complex with calc... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human TRPM4 channel in complex with calcium, decavanadate and ATP at 37 degrees Celsius | ||||||||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | ion channel / TRP channel / MEMBRANE PROTEIN | ||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Hu J / Lu W / Du J | ||||||||||||||||||||||||||||||
| Funding support |  United States, 9 items United States, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Physiological temperature drives TRPM4 ligand recognition and gating. Authors: Jinhong Hu / Sung Jin Park / Tyler Walter / Ian J Orozco / Garrett O'Dea / Xinyu Ye / Juan Du / Wei Lü /  Abstract: Temperature profoundly affects macromolecular function, particularly in proteins with temperature sensitivity. However, its impact is often overlooked in biophysical studies that are typically ...Temperature profoundly affects macromolecular function, particularly in proteins with temperature sensitivity. However, its impact is often overlooked in biophysical studies that are typically performed at non-physiological temperatures, potentially leading to inaccurate mechanistic and pharmacological insights. Here we demonstrate temperature-dependent changes in the structure and function of TRPM4, a temperature-sensitive Ca-activated ion channel. By studying TRPM4 prepared at physiological temperature using single-particle cryo-electron microscopy, we identified a 'warm' conformation that is distinct from those observed at lower temperatures. This conformation is driven by a temperature-dependent Ca-binding site in the intracellular domain, and is essential for TRPM4 function in physiological contexts. We demonstrated that ligands, exemplified by decavanadate (a positive modulator) and ATP (an inhibitor), bind to different locations of TRPM4 at physiological temperatures than at lower temperatures, and that these sites have bona fide functional relevance. We elucidated the TRPM4 gating mechanism by capturing structural snapshots of its different functional states at physiological temperatures, revealing the channel opening that is not observed at lower temperatures. Our study provides an example of temperature-dependent ligand recognition and modulation of an ion channel, underscoring the importance of studying macromolecules at physiological temperatures. It also provides a potential molecular framework for deciphering how thermosensitive TRPM channels perceive temperature changes. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44369.map.gz emd_44369.map.gz | 190.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44369-v30.xml emd-44369-v30.xml emd-44369.xml emd-44369.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44369.png emd_44369.png | 85.3 KB | ||
| Filedesc metadata |  emd-44369.cif.gz emd-44369.cif.gz | 5.5 KB | ||
| Others |  emd_44369_additional_1.map.gz emd_44369_additional_1.map.gz emd_44369_half_map_1.map.gz emd_44369_half_map_1.map.gz emd_44369_half_map_2.map.gz emd_44369_half_map_2.map.gz | 201.1 MB 199.5 MB 199.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44369 http://ftp.pdbj.org/pub/emdb/structures/EMD-44369 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44369 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44369 | HTTPS FTP |
-Validation report
| Summary document |  emd_44369_validation.pdf.gz emd_44369_validation.pdf.gz | 774 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44369_full_validation.pdf.gz emd_44369_full_validation.pdf.gz | 773.6 KB | Display | |
| Data in XML |  emd_44369_validation.xml.gz emd_44369_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_44369_validation.cif.gz emd_44369_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44369 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44369 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44369 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44369 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_44369.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44369.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_44369_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
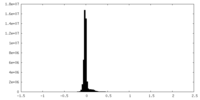
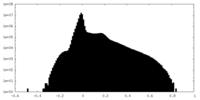
| Density Histograms |
-Half map: #1
| File | emd_44369_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
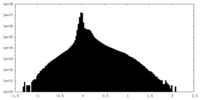
| Density Histograms |
-Half map: #2
| File | emd_44369_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
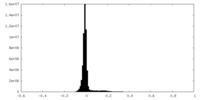
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the human TRPM4 channel in complex with calc...
| Entire | Name: Cryo-EM structure of the human TRPM4 channel in complex with calcium, decavanadate and ATP at 37 degrees Celsius |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the human TRPM4 channel in complex with calc...
| Supramolecule | Name: Cryo-EM structure of the human TRPM4 channel in complex with calcium, decavanadate and ATP at 37 degrees Celsius type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transient receptor potential cation channel subfamily M
| Macromolecule | Name: Transient receptor potential cation channel subfamily M type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: MVVPEKEQSW IPKIFKKKTC TTFIVDSTDP GGTLCQCGRP RTAHPAVAME DAFGAAVVTV WDSDAHTTEK PTDAYGELDF TGAGRKHSNF LRLSDRTDPA AVYSLVTRTW GFRAPNLVVS VLGGSGGPVL QTWLQDLLRR GLVRAAQSTG AWIVTGGLHT GIGRHVGVAV ...String: MVVPEKEQSW IPKIFKKKTC TTFIVDSTDP GGTLCQCGRP RTAHPAVAME DAFGAAVVTV WDSDAHTTEK PTDAYGELDF TGAGRKHSNF LRLSDRTDPA AVYSLVTRTW GFRAPNLVVS VLGGSGGPVL QTWLQDLLRR GLVRAAQSTG AWIVTGGLHT GIGRHVGVAV RDHQMASTGG TKVVAMGVAP WGVVRNRDTL INPKGSFPAR YRWRGDPEDG VQFPLDYNYS AFFLVDDGTH GCLGGENRFR LRLESYISQQ KTGVGGTGID IPVLLLLIDG DEKMLTRIEN ATQAQLPCLL VAGSGGAADC LAETLEDTLA PGSGGARQGE ARDRIRRFFP KGDLEVLQAQ VERIMTRKEL LTVYSSEDGS EEFETIVLKA LVKACGSSEA SAYLDELRLA VAWNRVDIAQ SELFRGDIQW RSFHLEASLM DALLNDRPEF VRLLISHGLS LGHFLTPMRL AQLYSAAPSN SLIRNLLDQA SHSAGTKAPA LKGGAAELRP PDVGHVLRML LGKMCAPRYP SGGAWDPHPG QGFGESMYLL SDKATSPLSL DAGLGQAPWS DLLLWALLLN RAQMAMYFWE MGSNAVSSAL GACLLLRVMA RLEPDAEEAA RRKDLAFKFE GMGVDLFGEC YRSSEVRAAR LLLRRCPLWG DATCLQLAMQ ADARAFFAQD GVQSLLTQKW WGDMASTTPI WALVLAFFCP PLIYTRLITF RKSEEEPTRE ELEFDMDSVI NGEGPVGTAD PAEKTPLGVP RQSGRPGCCG GRCGGRRCLR RWFHFWGAPV TIFMGNVVSY LLFLLLFSRV LLVDFQPAPP GSLELLLYFW AFTLLCEELR QGLSGGGGSL ASGGPGPGHA SLSQRLRLYL ADSWNQCDLV ALTCFLLGVG CRLTPGLYHL GRTVLCIDFM VFTVRLLHIF TVNKQLGPKI VIVSKMMKDV FFFLFFLGVW LVAYGVATEG LLRPRDSDFP SILRRVFYRP YLQIFGQIPQ EDMDVALMEH SNCSSEPGFW AHPPGAQAGT CVSQYANWLV VLLLVIFLLV ANILLVNLLI AMFSYTFGKV QGNSDLYWKA QRYRLIREFH SRPALAPPFI VISHLRLLLR QLCRRPRSPQ PSSPALEHFR VYLSKEAERK LLTWESVHKE NFLLARARDK RESDSERLKR TSQKVDLALK QLGHIREYEQ RLKVLEREVQ QCSRVLGWVA EALSRSALLP PGGPPPPDLP GSKD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 70000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





















 Z
Z Y
Y X
X