+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4410 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast RPA bound to ssDNA | |||||||||
 Map data Map data | 4.7 Angstrom map (sharpened) corresponding to Yeast RPA Trimerisation Core (Tri-C) bound by ssDNA. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / heterotrimer / DNA binding / OB-fold / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationheteroduplex formation / sporulation / DNA replication factor A complex / : / Removal of the Flap Intermediate / telomere maintenance via telomere lengthening / mitotic recombination / Translesion synthesis by REV1 / : / : ...heteroduplex formation / sporulation / DNA replication factor A complex / : / Removal of the Flap Intermediate / telomere maintenance via telomere lengthening / mitotic recombination / Translesion synthesis by REV1 / : / : / Activation of the pre-replicative complex / : / : / Activation of ATR in response to replication stress / single-stranded telomeric DNA binding / telomere maintenance via recombination / reciprocal meiotic recombination / telomeric DNA binding / Gap-filling DNA repair synthesis and ligation in TC-NER / DNA topological change / Dual incision in TC-NER / telomere maintenance via telomerase / telomere maintenance / condensed nuclear chromosome / meiotic cell cycle / nucleotide-excision repair / establishment of protein localization / double-strand break repair via homologous recombination / single-stranded DNA binding / site of double-strand break / double-stranded DNA binding / sequence-specific DNA binding / damaged DNA binding / DNA replication / chromosome, telomeric region / protein ubiquitination / DNA repair / mRNA binding / zinc ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
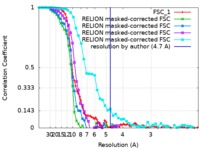
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Yates LA / Aramayo RJ | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Authors: Luke A Yates / Ricardo J Aramayo / Nilisha Pokhrel / Colleen C Caldwell / Joshua A Kaplan / Rajika L Perera / Maria Spies / Edwin Antony / Xiaodong Zhang /   Abstract: Replication Protein A (RPA), the major eukaryotic single stranded DNA-binding protein, binds to exposed ssDNA to protect it from nucleases, participates in a myriad of nucleic acid transactions and ...Replication Protein A (RPA), the major eukaryotic single stranded DNA-binding protein, binds to exposed ssDNA to protect it from nucleases, participates in a myriad of nucleic acid transactions and coordinates the recruitment of other important players. RPA is a heterotrimer and coats long stretches of single-stranded DNA (ssDNA). The precise molecular architecture of the RPA subunits and its DNA binding domains (DBDs) during assembly is poorly understood. Using cryo electron microscopy we obtained a 3D reconstruction of the RPA trimerisation core bound with ssDNA (∼55 kDa) at ∼4.7 Å resolution and a dimeric RPA assembly on ssDNA. FRET-based solution studies reveal dynamic rearrangements of DBDs during coordinated RPA binding and this activity is regulated by phosphorylation at S178 in RPA70. We present a structural model on how dynamic DBDs promote the cooperative assembly of multiple RPAs on long ssDNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4410.map.gz emd_4410.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4410-v30.xml emd-4410-v30.xml emd-4410.xml emd-4410.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4410_fsc_1.xml emd_4410_fsc_1.xml emd_4410_fsc_2.xml emd_4410_fsc_2.xml emd_4410_fsc_3.xml emd_4410_fsc_3.xml emd_4410_fsc_4.xml emd_4410_fsc_4.xml emd_4410_fsc_5.xml emd_4410_fsc_5.xml | 10 KB 3 KB 3 KB 3 KB 4.4 KB | Display Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_4410.png emd_4410.png | 95.6 KB | ||
| Filedesc metadata |  emd-4410.cif.gz emd-4410.cif.gz | 6.8 KB | ||
| Others |  emd_4410_additional_1.map.gz emd_4410_additional_1.map.gz emd_4410_additional_2.map.gz emd_4410_additional_2.map.gz emd_4410_additional_3.map.gz emd_4410_additional_3.map.gz emd_4410_additional_4.map.gz emd_4410_additional_4.map.gz | 49.4 MB 1.8 MB 1.8 MB 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4410 http://ftp.pdbj.org/pub/emdb/structures/EMD-4410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4410 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4410 | HTTPS FTP |
-Related structure data
| Related structure data |  6i52MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4410.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4410.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 4.7 Angstrom map (sharpened) corresponding to Yeast RPA Trimerisation Core (Tri-C) bound by ssDNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 7.5 Angstrom map (unsharpened) corresponding to yeast RPA dimer
| File | emd_4410_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 7.5 Angstrom map (unsharpened) corresponding to yeast RPA dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 7.7 Angstrom map (sharpened) corresponding to Yeast RPA...
| File | emd_4410_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 7.7 Angstrom map (sharpened) corresponding to Yeast RPA Trimerisation Core (Tri-C) bound by ssDNA with additional DNA-binding domain (DBD-B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 8.5 Angstrom map (sharpened) corresponding to Yeast RPA...
| File | emd_4410_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 8.5 Angstrom map (sharpened) corresponding to Yeast RPA Trimerisation Core (Tri-C) bound by ssDNA with additional DNA-binding domain (DBD-B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 10 Angstrom map (sharpened) corresponding to Yeast RPA...
| File | emd_4410_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 10 Angstrom map (sharpened) corresponding to Yeast RPA Trimerisation Core (Tri-C) bound by ssDNA with additional DNA-binding domain (DBD-B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Replication Protein A heterotrimeric complex with ssDNA
| Entire | Name: Replication Protein A heterotrimeric complex with ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: Replication Protein A heterotrimeric complex with ssDNA
| Supramolecule | Name: Replication Protein A heterotrimeric complex with ssDNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 120 KDa |
-Supramolecule #2: Replication Protein A heterotrimeric complex with ssDNA
| Supramolecule | Name: Replication Protein A heterotrimeric complex with ssDNA type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Location in cell: nucleus |
-Supramolecule #3: Replication Protein A heterotrimeric complex with ssDNA
| Supramolecule | Name: Replication Protein A heterotrimeric complex with ssDNA type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Replication factor A protein 3
| Macromolecule | Name: Replication factor A protein 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.827723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASETPRVDP TEISNVNAPV FRIIAQIKSQ PTESQLILQS PTISSKNGSE VEMITLNNIR VSMNKTFEID SWYEFVCRNN DDGELGFLI LDAVLCKFKE NEDLSLNGVV ALQRLCKKYP EIY UniProtKB: Replication factor A protein 3 |
-Macromolecule #2: Replication factor A protein 2
| Macromolecule | Name: Replication factor A protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.80987 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TNTRVNTLTP VTIKQILESK QDIQDGPFVS HNQELHHVCF VGVVRNITDH TANIFLTIED GTGQIEVRKW SEDAAQQFEI GGYVKVFGA LKEFGGKKNI QYAVIKPIDS FNEVLTHHLE VIKCHSIASG MMK UniProtKB: Replication factor A protein 2 |
-Macromolecule #3: Replication factor A protein 1
| Macromolecule | Name: Replication factor A protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.643072 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KFIAQRITIA RAQAENLGRS EKGDFFSVKA AISFLKVDNF AYPACSNENC NKKVLEQPDG TWRCEKCDTN NARPNWRYIL TISIIDETN QLWLTLFDDQ AKQLLGVDAN TLMSLKEEDP NEFTKITQSI QMNEYDFRIR AREDTYNDQS RIRYTVANLH S LNYRAEAD YLADELSKAL UniProtKB: Replication factor A protein 1 |
-Macromolecule #4: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP...
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.038899 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: glow-discharged grid, wait time 30 seconds, blotting time 1 second. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6i52: |
 Movie
Movie Controller
Controller



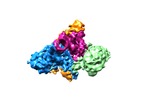

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)