[English] 日本語
 Yorodumi
Yorodumi- EMDB-43543: Cryo-EM structure of human ABC transporter (hABCC1) bound to cGAMP -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human ABC transporter (hABCC1) bound to cGAMP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cGAMP exporter / ABC transporter / multidrug-resistant protein / homodimer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsphingolipid translocation / ABC-type vitamin B12 transporter activity / Transport of RCbl within the body / cyclic nucleotide transport / carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / cobalamin transport / sphingolipid transporter activity / glutathione transmembrane transport / leukotriene transport ...sphingolipid translocation / ABC-type vitamin B12 transporter activity / Transport of RCbl within the body / cyclic nucleotide transport / carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / cobalamin transport / sphingolipid transporter activity / glutathione transmembrane transport / leukotriene transport / leukotriene metabolic process / ABC-type glutathione-S-conjugate transporter / ABC-type glutathione S-conjugate transporter activity / glutathione transmembrane transporter activity / Synthesis of Leukotrienes (LT) and Eoxins (EX) / sphingolipid biosynthetic process / : / Sphingolipid de novo biosynthesis / heme catabolic process / xenobiotic transport across blood-brain barrier / transepithelial transport / ATPase-coupled lipid transmembrane transporter activity / Paracetamol ADME / export across plasma membrane / NFE2L2 regulating MDR associated enzymes / ABC-type xenobiotic transporter / ABC-type xenobiotic transporter activity / Heme degradation / phospholipid translocation / efflux transmembrane transporter activity / lateral plasma membrane / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / xenobiotic transport / ABC-type transporter activity / transport across blood-brain barrier / xenobiotic metabolic process / basal plasma membrane / cell chemotaxis / Cytoprotection by HMOX1 / ABC-family proteins mediated transport / transmembrane transport / cellular response to amyloid-beta / positive regulation of inflammatory response / cellular response to oxidative stress / basolateral plasma membrane / apical plasma membrane / response to xenobiotic stimulus / ATP hydrolysis activity / extracellular exosome / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.54 Å | |||||||||
 Authors Authors | Shinde O / Li P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2025 Journal: Immunity / Year: 2025Title: Structures of ATP-binding cassette transporter ABCC1 reveal the molecular basis of cyclic dinucleotide cGAMP export. Authors: Omkar Shinde / Joshua A Boyer / Stephanie Cambier / Jordyn J VanPortfliet / Xuewu Sui / Gaya P Yadav / Elizabeth G Viverette / Mario J Borgnia / A Phillip West / Qi Zhang / Daniel B Stetson / Pingwei Li /  Abstract: Cyclic nucleotide GMP-AMP (cGAMP) plays a critical role in mediating the innate immune response through the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway. Recent ...Cyclic nucleotide GMP-AMP (cGAMP) plays a critical role in mediating the innate immune response through the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway. Recent studies showed that ATP-binding cassette subfamily C member 1 (ABCC1) is a cGAMP exporter. The exported cGAMP can be imported into uninfected cells to stimulate a STING-mediated innate immune response. However, the molecular basis of cGAMP export mediated by ABCC1 remains unclear. Here, we report the cryoelectron microscopy (cryo-EM) structures of human ABCC1 in a ligand-free state and a cGAMP-bound state. These structures reveal that ABCC1 forms a homodimer via its N-terminal transmembrane domain. The ligand-bound structure shows that cGAMP is recognized by a positively charged pocket. Mutagenesis and functional studies confirmed the roles of the ligand-binding pocket in cGAMP recognition and export. This study provides insights into the structure and function of ABCC1 as a cGAMP exporter and lays a foundation for future research targeting ABCC1 in infection and anti-cancer immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43543.map.gz emd_43543.map.gz | 136.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43543-v30.xml emd-43543-v30.xml emd-43543.xml emd-43543.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43543_fsc.xml emd_43543_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_43543.png emd_43543.png | 185.2 KB | ||
| Filedesc metadata |  emd-43543.cif.gz emd-43543.cif.gz | 7.2 KB | ||
| Others |  emd_43543_half_map_1.map.gz emd_43543_half_map_1.map.gz emd_43543_half_map_2.map.gz emd_43543_half_map_2.map.gz | 134.5 MB 134.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43543 http://ftp.pdbj.org/pub/emdb/structures/EMD-43543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43543 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43543 | HTTPS FTP |
-Related structure data
| Related structure data |  8vuxMC  8vt4C  8vvcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43543.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43543.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43543_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_43543_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homodimer of hABCC1 bound to cGAMP
| Entire | Name: Homodimer of hABCC1 bound to cGAMP |
|---|---|
| Components |
|
-Supramolecule #1: Homodimer of hABCC1 bound to cGAMP
| Supramolecule | Name: Homodimer of hABCC1 bound to cGAMP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 199 KDa |
-Macromolecule #1: Multidrug resistance-associated protein 1
| Macromolecule | Name: Multidrug resistance-associated protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 171.762953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALRGFCSAD GSDPLWDWNV TWNTSNPDFT KCFQNTVLVW VPCFYLWACF PFYFLYLSRH DRGYIQMTPL NKTKTALGFL LWIVCWADL FYSFWERSRG IFLAPVFLVS PTLLGITMLL ATFLIQLERR KGVQSSGIML TFWLVALVCA LAILRSKIMT A LKEDAQVD ...String: MALRGFCSAD GSDPLWDWNV TWNTSNPDFT KCFQNTVLVW VPCFYLWACF PFYFLYLSRH DRGYIQMTPL NKTKTALGFL LWIVCWADL FYSFWERSRG IFLAPVFLVS PTLLGITMLL ATFLIQLERR KGVQSSGIML TFWLVALVCA LAILRSKIMT A LKEDAQVD LFRDITFYVY FSLLLIQLVL SCFSDRSPLF SETIHDPNPC PESSASFLSR ITFWWITGLI VRGYRQPLEG SD LWSLNKE DTSEQVVPVL VKNWKKECAK TRKQPVKVVY SSKDPAQPKE SSKVDANEEV EALIVKSPQK EWNPSLFKVL YKT FGPYFL MSFFFKAIHD LMMFSGPQIL KLLIKFVNDT KAPDWQGYFY TVLLFVTACL QTLVLHQYFH ICFVSGMRIK TAVI GAVYR KALVITNSAR KSSTVGEIVN LMSVDAQRFM DLATYINMIW SAPLQVILAL YLLWLNLGPS VLAGVAVMVL MVPVN AVMA MKTKTYQVAH MKSKDNRIKL MNEILNGIKV LKLYAWELAF KDKVLAIRQE ELKVLKKSAY LSAVGTFTWV CTPFLV ALC TFAVYVTIDE NNILDAQTAF VSLALFNILR FPLNILPMVI SSIVQASVSL KRLRIFLSHE ELEPDSIERR PVKDGGG TN SITVRNATFT WARSDPPTLN GITFSIPEGA LVAVVGQVGC GKSSLLSALL AEMDKVEGHV AIKGSVAYVP QQAWIQND S LRENILFGCQ LEEPYYRSVI QACALLPDLE ILPSGDRTEI GEKGVNLSGG QKQRVSLARA VYSNADIYLF DDPLSAVDA HVGKHIFENV IGPKGMLKNK TRILVTHSMS YLPQVDVIIV MSGGKISEMG SYQELLARDG AFAEFLRTYA STEQEQDAEE NGVTGVSGP GKEAKQMENG MLVTDSAGKQ LQRQLSSSSS YSGDISRHHN STAELQKAEA KKEETWKLME ADKAQTGQVK L SVYWDYMK AIGLFISFLS IFLFMCNHVS ALASNYWLSL WTDDPIVNGT QEHTKVRLSV YGALGISQGI AVFGYSMAVS IG GILASRC LHVDLLHSIL RSPMSFFERT PSGNLVNRFS KELDTVDSMI PEVIKMFMGS LFNVIGACIV ILLATPIAAI IIP PLGLIY FFVQRFYVAS SRQLKRLESV SRSPVYSHFN ETLLGVSVIR AFEEQERFIH QSDLKVDENQ KAYYPSIVAN RWLA VRLEC VGNCIVLFAA LFAVISRHSL SAGLVGLSVS YSLQVTTYLN WLVRMSSEME TNIVAVERLK EYSETEKEAP WQIQE TAPP SSWPQVGRVE FRNYCLRYRE DLDFVLRHIN VTINGGEKVG IVGRTGAGKS SLTLGLFRIN ESAEGEIIID GINIAK IGL HDLRFKITII PQDPVLFSGS LRMNLDPFSQ YSDEEVWTSL ELAHLKDFVS ALPDKLDHEC AEGGENLSVG QRQLVCL AR ALLRKTKILV LDEATAAVDL ETDDLIQSTI RTQFEDCTVL TIAHRLNTIM DYTRVIVLDK GEIQEYGAPS DLLQQRGL F YSMAKDAGLV UniProtKB: Multidrug resistance-associated protein 1 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: cGAMP
| Macromolecule | Name: cGAMP / type: ligand / ID: 3 / Number of copies: 1 / Formula: 1SY |
|---|---|
| Molecular weight | Theoretical: 674.411 Da |
| Chemical component information | 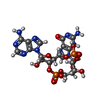 ChemComp-1SY: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































