[English] 日本語
 Yorodumi
Yorodumi- EMDB-43300: Raw consensus map of mouse RyR1 in complex with S100A1 (high-Ca2+... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Raw consensus map of mouse RyR1 in complex with S100A1 (high-Ca2+/CFF/ATP dataset) | |||||||||
 Map data Map data | Raw consensus map of mouse RyR1 in complex with S100A1 (high-Ca/CFF/ATP dataset) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Calcium / Ion Channel / MEMBRANE PROTEIN | |||||||||
| Biological species |  | |||||||||
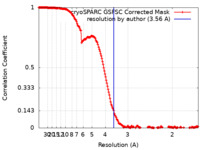
| Method | single particle reconstruction / cryo EM / Resolution: 3.56 Å | |||||||||
 Authors Authors | Weninger G / Marks AR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structural insights into the regulation of RyR1 by S100A1. Authors: Gunnar Weninger / Marco C Miotto / Carl Tchagou / Steven Reiken / Haikel Dridi / Sören Brandenburg / Gabriel C Riedemann / Qi Yuan / Yang Liu / Alexander Chang / Anetta Wronska / Stephan E ...Authors: Gunnar Weninger / Marco C Miotto / Carl Tchagou / Steven Reiken / Haikel Dridi / Sören Brandenburg / Gabriel C Riedemann / Qi Yuan / Yang Liu / Alexander Chang / Anetta Wronska / Stephan E Lehnart / Andrew R Marks /   Abstract: S100A1, a small homodimeric EF-hand Ca-binding protein (~21 kDa), plays an important regulatory role in Ca signaling pathways involved in various biological functions including Ca cycling and ...S100A1, a small homodimeric EF-hand Ca-binding protein (~21 kDa), plays an important regulatory role in Ca signaling pathways involved in various biological functions including Ca cycling and contractile performance in skeletal and cardiac myocytes. One key target of the S100A1 interactome is the ryanodine receptor (RyR), a huge homotetrameric Ca release channel (~2.3 MDa) of the sarcoplasmic reticulum. Here, we report cryoelectron microscopy structures of S100A1 bound to RyR1, the skeletal muscle isoform, in absence and presence of Ca. Ca-free apo-S100A1 binds beneath the bridging solenoid (BSol) and forms contacts with the junctional solenoid and the shell-core linker of RyR1. Upon Ca-binding, S100A1 undergoes a conformational change resulting in the exposure of the hydrophobic pocket known to serve as a major interaction site of S100A1. Through interactions of the hydrophobic pocket with RyR1, Ca-bound S100A1 intrudes deeper into the RyR1 structure beneath BSol than the apo-form and induces sideways motions of the C-terminal BSol region toward the adjacent RyR1 protomer resulting in tighter interprotomer contacts. Interestingly, the second hydrophobic pocket of the S100A1-dimer is largely exposed at the hydrophilic surface making it prone to interactions with the local environment, suggesting that S100A1 could be involved in forming larger heterocomplexes of RyRs with other protein partners. Since S100A1 interactions stabilizing BSol are implicated in the regulation of RyR-mediated Ca release, the characterization of the S100A1 binding site conserved between RyR isoforms may provide the structural basis for the development of therapeutic strategies regarding treatments of RyR-related disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43300.map.gz emd_43300.map.gz | 254.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43300-v30.xml emd-43300-v30.xml emd-43300.xml emd-43300.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43300_fsc.xml emd_43300_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_43300.png emd_43300.png | 106.9 KB | ||
| Filedesc metadata |  emd-43300.cif.gz emd-43300.cif.gz | 4.3 KB | ||
| Others |  emd_43300_half_map_1.map.gz emd_43300_half_map_1.map.gz emd_43300_half_map_2.map.gz emd_43300_half_map_2.map.gz | 474.3 MB 474.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43300 http://ftp.pdbj.org/pub/emdb/structures/EMD-43300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43300 | HTTPS FTP |
-Validation report
| Summary document |  emd_43300_validation.pdf.gz emd_43300_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43300_full_validation.pdf.gz emd_43300_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_43300_validation.xml.gz emd_43300_validation.xml.gz | 26.1 KB | Display | |
| Data in CIF |  emd_43300_validation.cif.gz emd_43300_validation.cif.gz | 34 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43300 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43300 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43300.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43300.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw consensus map of mouse RyR1 in complex with S100A1 (high-Ca/CFF/ATP dataset) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
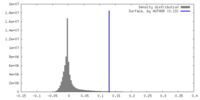
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map B
| File | emd_43300_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
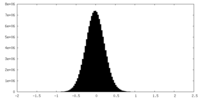
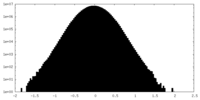
| Density Histograms |
-Half map: Half Map A
| File | emd_43300_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
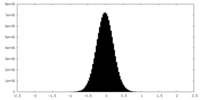
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of RyR1 with S100A1 and Calstabin-1 (high-Ca2+/CFF/ATP co...
| Entire | Name: Complex of RyR1 with S100A1 and Calstabin-1 (high-Ca2+/CFF/ATP condition) |
|---|---|
| Components |
|
-Supramolecule #1: Complex of RyR1 with S100A1 and Calstabin-1 (high-Ca2+/CFF/ATP co...
| Supramolecule | Name: Complex of RyR1 with S100A1 and Calstabin-1 (high-Ca2+/CFF/ATP condition) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: 0.25 mM free Ca2+; 5 mM Caffeine; 10 mM ATP |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.14 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||||||||
| Details | 0.15 mM S100A1-dimer |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 12555 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)