[English] 日本語
 Yorodumi
Yorodumi- EMDB-43290: Cryo-EM structure of Myxococcus xanthus EncA encapsulin shell loa... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Myxococcus xanthus EncA encapsulin shell loaded with EncD cargo | |||||||||
 Map data Map data | EncA-EncD encapsulin cryo-EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Encapsulin / EncA / EncD / flavin / flavin mononucleotide / iron / ferroxidase / ferritin / ferric reductase / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / iron ion transport / intracellular iron ion homeostasis / metal ion binding / Type 1 encapsulin shell protein EncA / Encapsulin nanocompartment cargo protein EncD Function and homology information Function and homology information | |||||||||
| Biological species |  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) | |||||||||
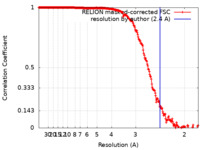
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Eren E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: encapsulin cargo protein EncD is a flavin-binding protein with ferric reductase activity. Authors: Elif Eren / Norman R Watts / James F Conway / Paul T Wingfield /  Abstract: Encapsulins are protein nanocompartments that regulate cellular metabolism in several bacteria and archaea. encapsulins protect the bacterial cells against oxidative stress by sequestering cytosolic ...Encapsulins are protein nanocompartments that regulate cellular metabolism in several bacteria and archaea. encapsulins protect the bacterial cells against oxidative stress by sequestering cytosolic iron. These encapsulins are formed by the shell protein EncA and three cargo proteins: EncB, EncC, and EncD. EncB and EncC form rotationally symmetric decamers with ferroxidase centers (FOCs) that oxidize Fe to Fe for iron storage in mineral form. However, the structure and function of the third cargo protein, EncD, have yet to be determined. Here, we report the x-ray crystal structure of EncD in complex with flavin mononucleotide. EncD forms an α-helical hairpin arranged as an antiparallel dimer, but unlike other flavin-binding proteins, it has no β-sheet, showing that EncD and its homologs represent a unique class of bacterial flavin-binding proteins. The cryo-EM structure of EncA-EncD encapsulins confirms that EncD binds to the interior of the EncA shell via its C-terminal targeting peptide. With only 100 amino acids, the EncD α-helical dimer forms the smallest flavin-binding domain observed to date. Unlike EncB and EncC, EncD lacks a FOC, and our biochemical results show that EncD instead is a NAD(P)H-dependent ferric reductase, indicating that the encapsulins act as an integrated system for iron homeostasis. Overall, this work contributes to our understanding of bacterial metabolism and could lead to the development of technologies for iron biomineralization and the production of iron-containing materials for the treatment of various diseases associated with oxidative stress. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43290.map.gz emd_43290.map.gz | 73.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43290-v30.xml emd-43290-v30.xml emd-43290.xml emd-43290.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43290_fsc.xml emd_43290_fsc.xml | 22.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_43290.png emd_43290.png | 255.8 KB | ||
| Filedesc metadata |  emd-43290.cif.gz emd-43290.cif.gz | 6.5 KB | ||
| Others |  emd_43290_half_map_1.map.gz emd_43290_half_map_1.map.gz emd_43290_half_map_2.map.gz emd_43290_half_map_2.map.gz | 471.3 MB 470.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43290 http://ftp.pdbj.org/pub/emdb/structures/EMD-43290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43290 | HTTPS FTP |
-Related structure data
| Related structure data |  8vjoMC  8vjnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43290.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43290.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EncA-EncD encapsulin cryo-EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EncA-EncD halfmap1
| File | emd_43290_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EncA-EncD halfmap1 | ||||||||||||
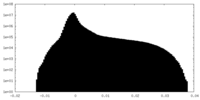
| Projections & Slices |
| ||||||||||||
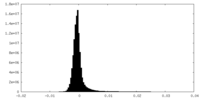
| Density Histograms |
-Half map: EncA-EncD halfmap2
| File | emd_43290_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EncA-EncD halfmap2 | ||||||||||||
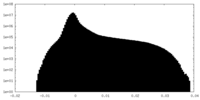
| Projections & Slices |
| ||||||||||||
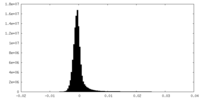
| Density Histograms |
- Sample components
Sample components
-Entire : Icosahedral encapsulin EncA particles in complex with cargo prote...
| Entire | Name: Icosahedral encapsulin EncA particles in complex with cargo protein EncD |
|---|---|
| Components |
|
-Supramolecule #1: Icosahedral encapsulin EncA particles in complex with cargo prote...
| Supramolecule | Name: Icosahedral encapsulin EncA particles in complex with cargo protein EncD type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
-Macromolecule #1: EncA
| Macromolecule | Name: EncA / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
| Molecular weight | Theoretical: 33.505074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHMPL EPHFMPDFLG HAENPLREEE WARLNETVIQ VARRSLVGRR ILDIYGPLGA GVQTVPYDEF QGVSPGAVDI VGEQETAMV FTDARKFKTI PIIYKDFLLH WRDIEAARTH NMPLDVSAAA GAAALCAQQE DELIFYGDAR LGYEGLMTAN G RLTVPLGD ...String: MHHHHHHMPL EPHFMPDFLG HAENPLREEE WARLNETVIQ VARRSLVGRR ILDIYGPLGA GVQTVPYDEF QGVSPGAVDI VGEQETAMV FTDARKFKTI PIIYKDFLLH WRDIEAARTH NMPLDVSAAA GAAALCAQQE DELIFYGDAR LGYEGLMTAN G RLTVPLGD WTSPGGGFQA IVEATRKLNE QGHFGPYAVV LSPRLYSQLH RIYEKTGVLE IETIRQLASD GVYQSNRLRG ES GVVVSTG RENMDLAVSM DMVAAYLGAS RMNHPFRVLE ALLLRIKHPD AICTLEGAGA TERR UniProtKB: Type 1 encapsulin shell protein EncA |
-Macromolecule #2: Encapsulin nanocompartment cargo protein EncD
| Macromolecule | Name: Encapsulin nanocompartment cargo protein EncD / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
| Molecular weight | Theoretical: 859.992 Da |
| Recombinant expression | Organism:  |
| Sequence | String: GLTVGSLRG UniProtKB: Encapsulin nanocompartment cargo protein EncD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.3 / Details: 20mM HEPES, 150mM NaCl |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)