+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | MicroED structure of SARS-CoV-2 main protease (MPro/3CLPro) with missing cone eliminated by suspended drop | ||||||||||||
 マップデータ マップデータ | 1.5 sigma | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Cysteine Protease / Viral Protease / Viral Assembly / VIRAL PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / 付加脱離酵素(リアーゼ); P-Oリアーゼ類; - / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / ISG15-specific peptidase activity / TRAF3-dependent IRF activation pathway ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / 付加脱離酵素(リアーゼ); P-Oリアーゼ類; - / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / ISG15-specific peptidase activity / TRAF3-dependent IRF activation pathway / Transcription of SARS-CoV-2 sgRNAs / Translation of Replicase and Assembly of the Replication Transcription Complex / snRNP Assembly / Replication of the SARS-CoV-2 genome / 加水分解酵素; エステル加水分解酵素; 5'-リン酸モノエステル産生エキソリボヌクレアーゼ / host cell endoplasmic reticulum-Golgi intermediate compartment / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / 5'-3' DNA helicase activity / 3'-5'-RNA exonuclease activity / host cell endosome / symbiont-mediated degradation of host mRNA / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / G-quadruplex RNA binding / symbiont-mediated suppression of host toll-like receptor signaling pathway / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / SARS-CoV-2 modulates host translation machinery / methyltransferase cap1 / host cell Golgi apparatus / symbiont-mediated suppression of host NF-kappaB cascade / symbiont-mediated perturbation of host ubiquitin-like protein modification / DNA helicase / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; システインプロテアーゼ / single-stranded RNA binding / host cell perinuclear region of cytoplasm / regulation of autophagy / viral protein processing / lyase activity / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / symbiont-mediated suppression of host gene expression / copper ion binding / viral translational frameshifting / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / lipid binding / DNA-templated transcription / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  | ||||||||||||
| 手法 | 電子線結晶学 / クライオ電子顕微鏡法 / 解像度: 2.15 Å | ||||||||||||
 データ登録者 データ登録者 | Bu G / Gillman C / Danelius E / Hattne J / Nannenga BL / Gonen T | ||||||||||||
| 資金援助 |  米国, 3件 米国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: J Struct Biol X / 年: 2024 ジャーナル: J Struct Biol X / 年: 2024タイトル: Eliminating the missing cone challenge through innovative approaches. 著者: Cody Gillman / Guanhong Bu / Emma Danelius / Johan Hattne / Brent L Nannenga / Tamir Gonen /  要旨: Microcrystal electron diffraction (MicroED) has emerged as a powerful technique for unraveling molecular structures from microcrystals too small for X-ray diffraction. However, a significant hurdle ...Microcrystal electron diffraction (MicroED) has emerged as a powerful technique for unraveling molecular structures from microcrystals too small for X-ray diffraction. However, a significant hurdle arises with plate-like crystals that consistently orient themselves flat on the electron microscopy grid. If the normal of the plate correlates with the axes of the crystal lattice, the crystal orientations accessible for measurement are restricted because the crystal cannot be arbitrarily rotated. This limits the information that can be acquired, resulting in a missing cone of information. We recently introduced a novel crystallization strategy called suspended drop crystallization and proposed that crystals in a suspended drop could effectively address the challenge of preferred crystal orientation. Here we demonstrate the success of the suspended drop approach in eliminating the missing cone in two samples that crystallize as thin plates: bovine liver catalase and the SARS‑CoV‑2 main protease (Mpro). This innovative solution proves indispensable for crystals exhibiting systematic preferred orientations, unlocking new possibilities for structure determination by MicroED. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_43144.map.gz emd_43144.map.gz | 1.8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-43144-v30.xml emd-43144-v30.xml emd-43144.xml emd-43144.xml | 14.7 KB 14.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_43144.png emd_43144.png | 126.1 KB | ||
| Filedesc metadata |  emd-43144.cif.gz emd-43144.cif.gz | 5.7 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43144 http://ftp.pdbj.org/pub/emdb/structures/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43144 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_43144_validation.pdf.gz emd_43144_validation.pdf.gz | 555.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_43144_full_validation.pdf.gz emd_43144_full_validation.pdf.gz | 554.8 KB | 表示 | |
| XML形式データ |  emd_43144_validation.xml.gz emd_43144_validation.xml.gz | 4.3 KB | 表示 | |
| CIF形式データ |  emd_43144_validation.cif.gz emd_43144_validation.cif.gz | 4.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43144 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43144 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43144 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8vd7MC  9bdjC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_43144.map.gz / 形式: CCP4 / 大きさ: 3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_43144.map.gz / 形式: CCP4 / 大きさ: 3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | 1.5 sigma | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 これらの図は立方格子座標系で作成されたものです | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X: 0.713 Å / Y: 0.694 Å / Z: 0.709 Å | ||||||||||||||||||||||||||||||||||||
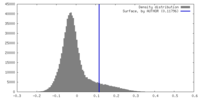
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 5 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : SARS-CoV-2 main protease (Mpro)
| 全体 | 名称: SARS-CoV-2 main protease (Mpro) |
|---|---|
| 要素 |
|
-超分子 #1: SARS-CoV-2 main protease (Mpro)
| 超分子 | 名称: SARS-CoV-2 main protease (Mpro) / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1 |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: 3C-like proteinase nsp5
| 分子 | 名称: 3C-like proteinase nsp5 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO / EC番号: SARS coronavirus main proteinase |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 33.825547 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: SGFRKMAFPS GKVEGCMVQV TCGTTTLNGL WLDDVVYCPR HVICTSEDML NPNYEDLLIR KSNHNFLVQA GNVQLRVIGH SMQNCVLKL KVDTANPKTP KYKFVRIQPG QTFSVLACYN GSPSGVYQCA MRPNFTIKGS FLNGSCGSVG FNIDYDCVSF C YMHHMELP ...文字列: SGFRKMAFPS GKVEGCMVQV TCGTTTLNGL WLDDVVYCPR HVICTSEDML NPNYEDLLIR KSNHNFLVQA GNVQLRVIGH SMQNCVLKL KVDTANPKTP KYKFVRIQPG QTFSVLACYN GSPSGVYQCA MRPNFTIKGS FLNGSCGSVG FNIDYDCVSF C YMHHMELP TGVHAGTDLE GNFYGPFVDR QTAQAAGTDT TITVNVLAWL YAAVINGDRW FLNRFTTTLN DFNLVAMKYN YE PLTQDHV DILGPLSAQT GIAVLDMCAS LKELLQNGMN GRTILGSALL EDEFTPFDVV RQCSGVTFQ UniProtKB: Replicase polyprotein 1ab |
-分子 #2: CHLORIDE ION
| 分子 | 名称: CHLORIDE ION / タイプ: ligand / ID: 2 / コピー数: 1 / 式: CL |
|---|---|
| 分子量 | 理論値: 35.453 Da |
-分子 #3: water
| 分子 | 名称: water / タイプ: ligand / ID: 3 / コピー数: 6 / 式: HOH |
|---|---|
| 分子量 | 理論値: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 電子線結晶学 |
| 試料の集合状態 | 3D array |
- 試料調製
試料調製
| 緩衝液 | pH: 6.5 / 詳細: 0.1 M MES pH 6.5, 20% PEG 3350, 5% DMSO. |
|---|---|
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 95 % / チャンバー内温度: 293 K |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 温度 | 最低: 75.0 K / 最高: 85.0 K |
| 撮影 | フィルム・検出器のモデル: FEI FALCON IV (4k x 4k) 回折像の数: 420 / 平均露光時間: 1.0 sec. / 平均電子線量: 0.0025 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: DIFFRACTION / 最大 デフォーカス(公称値): 0.0 µm / 最小 デフォーカス(公称値): 0.0 µm / カメラ長: 2480 mm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN / 傾斜角度: -40.0, 70.0 |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 2.15 Å / 解像度の算出法: DIFFRACTION PATTERN/LAYERLINES | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystallography statistics | Number intensities measured: 123734 / Number structure factors: 14825 / Fourier space coverage: 95.5 / R merge: 25.6 / Overall phase residual: 0 / Phase error rejection criteria: none / High resolution: 2.15 Å 殻:
|
-原子モデル構築 1
| 精密化 | プロトコル: OTHER 当てはまり具合の基準: Rwork/Rfree gap of less than 5% |
|---|---|
| 得られたモデル |  PDB-8vd7: |
 ムービー
ムービー コントローラー
コントローラー













 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)