[English] 日本語
 Yorodumi
Yorodumi- EMDB-43102: Structure of the E. coli clamp loader bound to the beta clamp in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the E. coli clamp loader bound to the beta clamp in a Open-RNAp/t conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial Clamp Loader Complex / REPLICATION / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA polymerase III, clamp loader complex / Hda-beta clamp complex / bacterial-type DNA replication / replication inhibiting complex / DNA clamp loader activity / DNA polymerase III complex / replisome / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / DNA polymerase processivity factor activity ...DNA polymerase III, clamp loader complex / Hda-beta clamp complex / bacterial-type DNA replication / replication inhibiting complex / DNA clamp loader activity / DNA polymerase III complex / replisome / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / DNA polymerase processivity factor activity / error-prone translesion synthesis / 3'-5' exonuclease activity / negative regulation of DNA-templated DNA replication initiation / ribonucleoside triphosphate phosphatase activity / DNA-templated DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / viral translational frameshifting / DNA damage response / protein homodimerization activity / ATP hydrolysis activity / DNA binding / ATP binding / metal ion binding / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
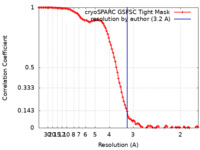
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Landeck JT / Kelch BA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Differences between bacteria and eukaryotes in clamp loader mechanism, a conserved process underlying DNA replication. Authors: Jacob T Landeck / Joshua Pajak / Emily K Norman / Emma L Sedivy / Brian A Kelch /  Abstract: Clamp loaders are pentameric ATPases that place circular sliding clamps onto DNA, where they function in DNA replication and genome integrity. The central activity of a clamp loader is the opening of ...Clamp loaders are pentameric ATPases that place circular sliding clamps onto DNA, where they function in DNA replication and genome integrity. The central activity of a clamp loader is the opening of the ring-shaped sliding clamp and the subsequent binding to primer-template (p/t)-junctions. The general architecture of clamp loaders is conserved across all life, suggesting that their mechanism is retained. Recent structural studies of the eukaryotic clamp loader replication factor C (RFC) revealed that it functions using a crab-claw mechanism, where clamp opening is coupled to a massive conformational change in the loader. Here we investigate the clamp loading mechanism of the Escherichia coli clamp loader at high resolution using cryo-electron microscopy. We find that the E. coli clamp loader opens the clamp using a crab-claw motion at a single pivot point, whereas the eukaryotic RFC loader uses motions distributed across the complex. Furthermore, we find clamp opening occurs in multiple steps, starting with a partly open state with a spiral conformation, and proceeding to a wide open clamp in a surprising planar geometry. Finally, our structures in the presence of p/t-junctions illustrate how the clamp closes around p/t-junctions and how the clamp loader initiates release from the loaded clamp. Our results reveal mechanistic distinctions in a macromolecular machine that is conserved across all domains of life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43102.map.gz emd_43102.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43102-v30.xml emd-43102-v30.xml emd-43102.xml emd-43102.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43102_fsc.xml emd_43102_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_43102.png emd_43102.png | 64.7 KB | ||
| Filedesc metadata |  emd-43102.cif.gz emd-43102.cif.gz | 7.7 KB | ||
| Others |  emd_43102_half_map_1.map.gz emd_43102_half_map_1.map.gz emd_43102_half_map_2.map.gz emd_43102_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43102 http://ftp.pdbj.org/pub/emdb/structures/EMD-43102 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43102 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43102 | HTTPS FTP |
-Related structure data
| Related structure data |  8vatMC  8valC  8vamC  8vanC  8vapC  8vaqC  8varC  8vasC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43102.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43102.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_43102_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43102_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Structure of the E. coli clamp loader bound to the beta clamp in ...
+Supramolecule #1: Structure of the E. coli clamp loader bound to the beta clamp in ...
+Macromolecule #1: DNA polymerase III subunit delta
+Macromolecule #2: DNA polymerase III subunit tau
+Macromolecule #3: DNA polymerase III subunit delta'
+Macromolecule #4: Beta sliding clamp
+Macromolecule #5: RNA (5'-R(P*AP*GP*UP*GP*GP*UP*GP*UP*CP*UP*G)-3')
+Macromolecule #6: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*CP*AP*GP*AP*CP*AP*CP*CP*AP*CP*TP...
+Macromolecule #7: ZINC ION
+Macromolecule #8: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #9: BERYLLIUM TRIFLUORIDE ION
+Macromolecule #10: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R2/2 / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 3132 / Average electron dose: 45.2 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.1 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 92.3 | ||||||
| Output model |  PDB-8vat: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)