[English] 日本語
 Yorodumi
Yorodumi- EMDB-43084: S. aureus TarL H300N in complex with CDP-ribitol (two tetramers w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. aureus TarL H300N in complex with CDP-ribitol (two tetramers with CHAPS micelle) | |||||||||||||||
 Map data Map data | Sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | glycosyltransferase / polymerase / monotopic / amphipathic / TRANSFERASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCDP-ribitol ribitolphosphotransferase / CDP-ribitol ribitolphosphotransferase activity / CDP-glycerol glycerophosphotransferase activity / teichoic acid biosynthetic process / cell wall organization / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Li FKK / Strynadka NCJ / Worrall LJ | |||||||||||||||
| Funding support |  Canada, Canada,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Cryo-EM analysis of TarL, a polymerase in wall teichoic acid biogenesis central to virulence and antibiotic resistance. Authors: Franco K K Li / Liam J Worrall / Robert T Gale / Eric D Brown / Natalie C J Strynadka /  Abstract: Wall teichoic acid (WTA), a covalent adduct of Gram-positive bacterial cell wall peptidoglycan, contributes directly to virulence and antibiotic resistance in pathogenic species. Polymerization of ...Wall teichoic acid (WTA), a covalent adduct of Gram-positive bacterial cell wall peptidoglycan, contributes directly to virulence and antibiotic resistance in pathogenic species. Polymerization of the WTA ribitol-phosphate chain is catalyzed by TarL, a member of the largely uncharacterized TagF-like family of membrane-associated enzymes. We report the cryo-electron microscopy structure of TarL, showing a tetramer that forms an extensive membrane-binding platform of monotopic helices. TarL is composed of an amino-terminal immunoglobulin-like domain and a carboxyl-terminal glycosyltransferase-B domain for ribitol-phosphate polymerization. The active site of the latter is complexed to donor substrate cytidine diphosphate-ribitol, providing mechanistic insights into the catalyzed phosphotransfer reaction. Furthermore, the active site is surrounded by electropositive residues that serve to retain the lipid-linked acceptor for polymerization. Our data advance general insight into the architecture and membrane association of the still poorly characterized monotopic membrane protein class and present molecular details of ribitol-phosphate polymerization that may aid in the design of new antimicrobials. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43084.map.gz emd_43084.map.gz | 56.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43084-v30.xml emd-43084-v30.xml emd-43084.xml emd-43084.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43084.png emd_43084.png | 97.9 KB | ||
| Masks |  emd_43084_msk_1.map emd_43084_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43084.cif.gz emd-43084.cif.gz | 5.4 KB | ||
| Others |  emd_43084_half_map_1.map.gz emd_43084_half_map_1.map.gz emd_43084_half_map_2.map.gz emd_43084_half_map_2.map.gz | 58.8 MB 58.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43084 http://ftp.pdbj.org/pub/emdb/structures/EMD-43084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43084 | HTTPS FTP |
-Validation report
| Summary document |  emd_43084_validation.pdf.gz emd_43084_validation.pdf.gz | 707.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43084_full_validation.pdf.gz emd_43084_full_validation.pdf.gz | 707.3 KB | Display | |
| Data in XML |  emd_43084_validation.xml.gz emd_43084_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_43084_validation.cif.gz emd_43084_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43084 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43084 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43084 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43084 | HTTPS FTP |
-Related structure data
| Related structure data |  8v33C  8v34C  8va1C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43084.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43084.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32375 Å | ||||||||||||||||||||||||||||||||||||
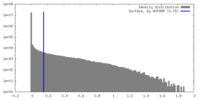
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43084_msk_1.map emd_43084_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
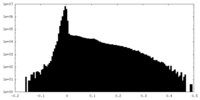
| Density Histograms |
-Half map: Half map B
| File | emd_43084_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_43084_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : S. aureus TarL H300N with CDP-ribitol and CHAPS
| Entire | Name: S. aureus TarL H300N with CDP-ribitol and CHAPS |
|---|---|
| Components |
|
-Supramolecule #1: S. aureus TarL H300N with CDP-ribitol and CHAPS
| Supramolecule | Name: S. aureus TarL H300N with CDP-ribitol and CHAPS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Two tetramers with CHAPS micelle |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: TarL H300N
| Macromolecule | Name: TarL H300N / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MVKSKIYIDK IYWERVQLFV EGHSENLDL EDSNFVLRNL T ETRTMKAN DVKIDGNQFV CR FNVAILD NGYYLPEDKY LLV NEQELD YIAQLNPDVI NDAY QNLKP EQEEEYNELE TQNGK INFL LQTYLKEFRK GGISKK TVY TVTPEISSDV NEFVLDV VV ...String: MVKSKIYIDK IYWERVQLFV EGHSENLDL EDSNFVLRNL T ETRTMKAN DVKIDGNQFV CR FNVAILD NGYYLPEDKY LLV NEQELD YIAQLNPDVI NDAY QNLKP EQEEEYNELE TQNGK INFL LQTYLKEFRK GGISKK TVY TVTPEISSDV NEFVLDV VV TTPEVKSIYI VRKYKELR K YFRKQSFNTR QFIFKAIFN TTKFFHLKKG NTVLFTSDSR PTMSGNFEY IYNEMLRQNL D KKYDIHTV FKANITDRRG II DKFRLPY LLGKADYIFV DDF HPLIYT VRFRRSQEVI QVWN AVGAF KTVGFSRTGK KGGPF IDSL NHRSYTKAYV SSETDI PFY AEAFGIKEKN VVPTGVP RT DVLFDEAYAT QIKQEMED E LPIIKGKKVI LFAPTFRGS GHGTAHYPFF KIDFERLARY CEKNNAVVL FKMHPFVKNR L NIADKHKQ YFVDVSDFRE VN DILFITD LLISDYSSLI YEY AVFKKP MIFYAFDLED YITT RDFYE PYESFVPGKI VQSFD ALMD ALDNEDYEGE KVIPFL DKH FKYQDGRSSE RLVRNLF GS KLVPRGSAAA ALEHHHHH H HH UniProtKB: Teichoic acid ribitol-phosphate polymerase TarL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 19380 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D4 (2x4 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 1341400 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)