[English] 日本語
 Yorodumi
Yorodumi- EMDB-43083: S. aureus TarL H300N in complex with CDP-ribitol (single tetramer) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. aureus TarL H300N in complex with CDP-ribitol (single tetramer) | |||||||||||||||
 Map data Map data | Sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | glycosyltransferase / polymerase / monotopic / amphipathic / TRANSFERASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCDP-ribitol ribitolphosphotransferase / CDP-ribitol ribitolphosphotransferase activity / CDP-glycerol glycerophosphotransferase activity / teichoic acid biosynthetic process / cell wall organization / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Li FKK / Worrall LJ / Strynadka NCJ | |||||||||||||||
| Funding support |  Canada, Canada,  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Cryo-EM analysis of TarL, a polymerase in wall teichoic acid biogenesis central to virulence and antibiotic resistance. Authors: Franco K K Li / Liam J Worrall / Robert T Gale / Eric D Brown / Natalie C J Strynadka /  Abstract: Wall teichoic acid (WTA), a covalent adduct of Gram-positive bacterial cell wall peptidoglycan, contributes directly to virulence and antibiotic resistance in pathogenic species. Polymerization of ...Wall teichoic acid (WTA), a covalent adduct of Gram-positive bacterial cell wall peptidoglycan, contributes directly to virulence and antibiotic resistance in pathogenic species. Polymerization of the WTA ribitol-phosphate chain is catalyzed by TarL, a member of the largely uncharacterized TagF-like family of membrane-associated enzymes. We report the cryo-electron microscopy structure of TarL, showing a tetramer that forms an extensive membrane-binding platform of monotopic helices. TarL is composed of an amino-terminal immunoglobulin-like domain and a carboxyl-terminal glycosyltransferase-B domain for ribitol-phosphate polymerization. The active site of the latter is complexed to donor substrate cytidine diphosphate-ribitol, providing mechanistic insights into the catalyzed phosphotransfer reaction. Furthermore, the active site is surrounded by electropositive residues that serve to retain the lipid-linked acceptor for polymerization. Our data advance general insight into the architecture and membrane association of the still poorly characterized monotopic membrane protein class and present molecular details of ribitol-phosphate polymerization that may aid in the design of new antimicrobials. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43083.map.gz emd_43083.map.gz | 57.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43083-v30.xml emd-43083-v30.xml emd-43083.xml emd-43083.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43083.png emd_43083.png | 227.4 KB | ||
| Masks |  emd_43083_msk_1.map emd_43083_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43083.cif.gz emd-43083.cif.gz | 6.4 KB | ||
| Others |  emd_43083_half_map_1.map.gz emd_43083_half_map_1.map.gz emd_43083_half_map_2.map.gz emd_43083_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43083 http://ftp.pdbj.org/pub/emdb/structures/EMD-43083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43083 | HTTPS FTP |
-Related structure data
| Related structure data |  8va1MC  8v33C  8v34C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43083.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43083.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32375 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43083_msk_1.map emd_43083_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_43083_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_43083_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : S. aureus TarL H300N with CDP-ribitol and CHAPS
| Entire | Name: S. aureus TarL H300N with CDP-ribitol and CHAPS |
|---|---|
| Components |
|
-Supramolecule #1: S. aureus TarL H300N with CDP-ribitol and CHAPS
| Supramolecule | Name: S. aureus TarL H300N with CDP-ribitol and CHAPS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Teichoic acid ribitol-phosphate polymerase TarL
| Macromolecule | Name: Teichoic acid ribitol-phosphate polymerase TarL / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: CDP-ribitol ribitolphosphotransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.510703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVKSKIYIDK IYWERVQLFV EGHSENLDLE DSNFVLRNLT ETRTMKANDV KIDGNQFVCR FNVAILDNGY YLPEDKYLLV NEQELDYIA QLNPDVINDA YQNLKPEQEE EYNELETQNG KINFLLQTYL KEFRKGGISK KTVYTVTPEI SSDVNEFVLD V VVTTPEVK ...String: MVKSKIYIDK IYWERVQLFV EGHSENLDLE DSNFVLRNLT ETRTMKANDV KIDGNQFVCR FNVAILDNGY YLPEDKYLLV NEQELDYIA QLNPDVINDA YQNLKPEQEE EYNELETQNG KINFLLQTYL KEFRKGGISK KTVYTVTPEI SSDVNEFVLD V VVTTPEVK SIYIVRKYKE LRKYFRKQSF NTRQFIFKAI FNTTKFFHLK KGNTVLFTSD SRPTMSGNFE YIYNEMLRQN LD KKYDIHT VFKANITDRR GIIDKFRLPY LLGKADYIFV DDFHPLIYTV RFRRSQEVIQ VWNAVGAFKT VGFSRTGKKG GPF IDSLNH RSYTKAYVSS ETDIPFYAEA FGIKEKNVVP TGVPRTDVLF DEAYATQIKQ EMEDELPIIK GKKVILFAPT FRGS GHGTA HYPFFKIDFE RLARYCEKNN AVVLFKMHPF VKNRLNIADK HKQYFVDVSD FREVNDILFI TDLLISDYSS LIYEY AVFK KPMIFYAFDL EDYITTRDFY EPYESFVPGK IVQSFDALMD ALDNEDYEGE KVIPFLDKHF KYQDGRSSER LVRNLF GSK LVPRGSAAAA LEHHHHHHHH UniProtKB: Teichoic acid ribitol-phosphate polymerase TarL |
-Macromolecule #2: CDP-ribitol
| Macromolecule | Name: CDP-ribitol / type: ligand / ID: 2 / Number of copies: 4 / Formula: V2V |
|---|---|
| Molecular weight | Theoretical: 537.307 Da |
| Chemical component information | 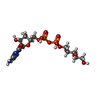 ChemComp-V2V: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 19380 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 190753 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)