[English] 日本語
 Yorodumi
Yorodumi- EMDB-43023: 60S ribosome biogenesis intermediate (Dbp10 catalytic structure -... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Low-pass filtered locally refined map) | ||||||||||||
 Map data Map data | Low pass filtered map of locally refined map (Dbp10 catalytic state) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | RNA Binding / RNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmaturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / preribosome, large subunit precursor / assembly of large subunit precursor of preribosome / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA processing / RNA helicase activity / RNA helicase / nucleolus / ATP hydrolysis activity / RNA binding ...maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / preribosome, large subunit precursor / assembly of large subunit precursor of preribosome / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / rRNA processing / RNA helicase activity / RNA helicase / nucleolus / ATP hydrolysis activity / RNA binding / ATP binding / identical protein binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||
 Authors Authors | Cruz VE / Weirich CS / Peddada N / Erzberger JP | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis. Authors: Victor E Cruz / Christine S Weirich / Nagesh Peddada / Jan P Erzberger /  Abstract: DEAD-box ATPases play crucial roles in guiding rRNA restructuring events during the biogenesis of large (60S) ribosomal subunits, but their precise molecular functions are currently unknown. In this ...DEAD-box ATPases play crucial roles in guiding rRNA restructuring events during the biogenesis of large (60S) ribosomal subunits, but their precise molecular functions are currently unknown. In this study, we present cryo-EM reconstructions of nucleolar pre-60S intermediates that reveal an unexpected, alternate secondary structure within the nascent peptidyl-transferase-center (PTC). Our analysis of three sequential nucleolar pre-60S intermediates reveals that the DEAD-box ATPase Dbp10/DDX54 remodels this alternate base pairing and enables the formation of the rRNA junction that anchors the mature form of the universally conserved PTC A-loop. Post-catalysis, Dbp10 captures rRNA helix H61, initiating the concerted exchange of biogenesis factors during late nucleolar 60S maturation. Our findings show that Dbp10 activity is essential for the formation of the ribosome active site and reveal how this function is integrated with subsequent assembly steps to drive the biogenesis of the large ribosomal subunit. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43023.map.gz emd_43023.map.gz | 220.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43023-v30.xml emd-43023-v30.xml emd-43023.xml emd-43023.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43023.png emd_43023.png | 37.3 KB | ||
| Filedesc metadata |  emd-43023.cif.gz emd-43023.cif.gz | 6.1 KB | ||
| Others |  emd_43023_half_map_1.map.gz emd_43023_half_map_1.map.gz emd_43023_half_map_2.map.gz emd_43023_half_map_2.map.gz | 193.8 MB 193.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43023 http://ftp.pdbj.org/pub/emdb/structures/EMD-43023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43023 | HTTPS FTP |
-Validation report
| Summary document |  emd_43023_validation.pdf.gz emd_43023_validation.pdf.gz | 853.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43023_full_validation.pdf.gz emd_43023_full_validation.pdf.gz | 852.8 KB | Display | |
| Data in XML |  emd_43023_validation.xml.gz emd_43023_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_43023_validation.cif.gz emd_43023_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43023 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43023 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43023 | HTTPS FTP |
-Related structure data
| Related structure data |  8v85MC  8v83C  8v84C  8v87C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43023.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43023.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Low pass filtered map of locally refined map (Dbp10 catalytic state) | ||||||||||||||||||||||||||||||||||||
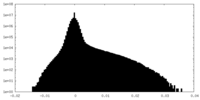
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map A
| File | emd_43023_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
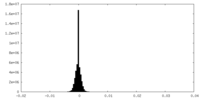
| Density Histograms |
-Half map: Half Map B
| File | emd_43023_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
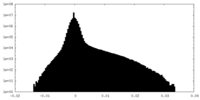
| Projections & Slices |
| ||||||||||||
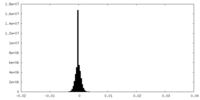
| Density Histograms |
- Sample components
Sample components
-Entire : 60S ribosome biogenesis intermediate
| Entire | Name: 60S ribosome biogenesis intermediate |
|---|---|
| Components |
|
-Supramolecule #1: 60S ribosome biogenesis intermediate
| Supramolecule | Name: 60S ribosome biogenesis intermediate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent RNA helicase DBP10
| Macromolecule | Name: ATP-dependent RNA helicase DBP10 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 113.126469 KDa |
| Sequence | String: MAGVQKRKRD LEDQDDNGSE EDDIAFDIAN EIALNDSESD ANDSDSEVEA DYGPNDVQDV IEYSSDEEEG VNNKKKAENK DIKKKKNSK KEIAAFPMLE MSDDENNASG KTQTGDDEDD VNEYFSTNNL EKTKHKKGSF PSFGLSKIVL NNIKRKGFRQ P TPIQRKTI ...String: MAGVQKRKRD LEDQDDNGSE EDDIAFDIAN EIALNDSESD ANDSDSEVEA DYGPNDVQDV IEYSSDEEEG VNNKKKAENK DIKKKKNSK KEIAAFPMLE MSDDENNASG KTQTGDDEDD VNEYFSTNNL EKTKHKKGSF PSFGLSKIVL NNIKRKGFRQ P TPIQRKTI PLILQSRDIV GMARTGSGKT AAFILPMVEK LKSHSGKIGA RAVILSPSRE LAMQTFNVFK DFARGTELRS VL LTGGDSL EEQFGMMMTN PDVIIATPGR FLHLKVEMNL DLKSVEYVVF DEADRLFEMG FQEQLNELLA SLPTTRQTLL FSA TLPNSL VDFVKAGLVN PVLVRLDAET KVSENLEMLF LSSKNADREA NLLYILQEII KIPLATSEQL QKLQNSNNEA DSDS DDEND RQKKRRNFKK EKFRKQKMPA ANELPSEKAT ILFVPTRHHV EYISQLLRDC GYLISYIYGT LDQHARKRQL YNFRA GLTS ILVVTDVAAR GVDIPMLANV INYTLPGSSK IFVHRVGRTA RAGNKGWAYS IVAENELPYL LDLELFLGKK ILLTPM YDS LVDVMKKRWI DEGKPEYQFQ PPKLSYTKRL VLGSCPRLDV EGLGDLYKNL MSSNFDLQLA KKTAMKAEKL YYRTRTS AS PESLKRSKEI ISSGWDAQNA FFGKNEEKEK LDFLAKLQNR RNKETVFEFT RNPDDEMAVF MKRRRKQLAP IQRKATER R ELLEKERMAG LSHSIEDEIL KGDDGETGYT VSEDALKEFE DADQLLEAQE NENKKKKKPK SFKDPTFFLS HYAPAGDIQ DKQLQITNGF ANDAAQAAYD LNSDDKVQVH KQTATVKWDK KRKKYVNTQG IDNKKYIIGE SGQKIAASFR SGRFDDWSKA RNLKPLKVG SRETSIPSNL LEDPSQGPAA NGRTVRGKFK HKQMKAPKMP DKHRDNYYSQ KKKVEKALQS GISVKGYNNA P GLRSELKS TEQIRKDRII AEKKRAKNAR PSKKRKF UniProtKB: ATP-dependent RNA helicase DBP10 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 39.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 4) Details: High-resolution map was low pass filtered to 6A to allow domain docking Number images used: 195000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4) |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)