+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4273 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fo conformation 1 | |||||||||
 Map data Map data | Chloroplast Fo and peripheral stalk conformation 1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
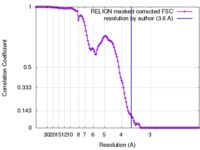
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Hahn A / Vonck J / Mills DJ / Kuehlbrandt W / Meier T | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure, mechanism, and regulation of the chloroplast ATP synthase. Authors: Alexander Hahn / Janet Vonck / Deryck J Mills / Thomas Meier / Werner Kühlbrandt /  Abstract: The chloroplast adenosine triphosphate (ATP) synthase uses the electrochemical proton gradient generated by photosynthesis to produce ATP, the energy currency of all cells. Protons conducted through ...The chloroplast adenosine triphosphate (ATP) synthase uses the electrochemical proton gradient generated by photosynthesis to produce ATP, the energy currency of all cells. Protons conducted through the membrane-embedded F motor drive ATP synthesis in the F head by rotary catalysis. We determined the high-resolution structure of the complete cFF complex by cryo-electron microscopy, resolving side chains of all 26 protein subunits, the five nucleotides in the F head, and the proton pathway to and from the rotor ring. The flexible peripheral stalk redistributes differences in torsional energy across three unequal steps in the rotation cycle. Plant ATP synthase is autoinhibited by a β-hairpin redox switch in subunit γ that blocks rotation in the dark. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4273.map.gz emd_4273.map.gz | 153 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4273-v30.xml emd-4273-v30.xml emd-4273.xml emd-4273.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4273_fsc.xml emd_4273_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4273.png emd_4273.png | 83.6 KB | ||
| Masks |  emd_4273_msk_1.map emd_4273_msk_1.map | 163.6 MB |  Mask map Mask map | |
| Others |  emd_4273_half_map_1.map.gz emd_4273_half_map_1.map.gz emd_4273_half_map_2.map.gz emd_4273_half_map_2.map.gz | 6.8 MB 129.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4273 http://ftp.pdbj.org/pub/emdb/structures/EMD-4273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4273 | HTTPS FTP |
-Related structure data
| Related structure data |  4270C  4271C  4272C  6fkfC  6fkhC  6fkiC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4273.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4273.map.gz / Format: CCP4 / Size: 163.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chloroplast Fo and peripheral stalk conformation 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.053 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4273_msk_1.map emd_4273_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_4273_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4273_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chloroplast Fo
| Entire | Name: Chloroplast Fo |
|---|---|
| Components |
|
-Supramolecule #1: Chloroplast Fo
| Supramolecule | Name: Chloroplast Fo / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#13 / Details: masked and realigned Fo from F1Fo complex. |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Molecular weight | Theoretical: 600 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number real images: 6254 / Average exposure time: 62.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.5 µm / Calibrated magnification: 132953 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | real space refinement as described for EMD-4270 |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)