[English] 日本語
 Yorodumi
Yorodumi- EMDB-42685: Structure of Ran bound to RCC1 and the nucleosome core particle (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Ran bound to RCC1 and the nucleosome core particle (composite map) | |||||||||
 Map data Map data | Composite map of the Ran-RCC1-NCP complex. The Ran-RCC1 portion was taken from a focus-refined Ran-RCC1 map. The NCP portion was taken from a 1-side bound Ran-RCC1-NCP map. Both local-sharpened. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GTPase / Guanine nucleotide exchange factor / chromatin-binding / NUCLEAR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHDMs demethylate histones / PKMTs methylate histone lysines / Condensation of Prophase Chromosomes / SUMOylation of chromatin organization proteins / Metalloprotease DUBs / E3 ubiquitin ligases ubiquitinate target proteins / RCAF complex / RMTs methylate histone arginines / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / polytene chromosome band ...HDMs demethylate histones / PKMTs methylate histone lysines / Condensation of Prophase Chromosomes / SUMOylation of chromatin organization proteins / Metalloprotease DUBs / E3 ubiquitin ligases ubiquitinate target proteins / RCAF complex / RMTs methylate histone arginines / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / polytene chromosome band / SIRT1 negatively regulates rRNA expression / NoRC negatively regulates rRNA expression / Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 / Formation of the beta-catenin:TCF transactivating complex / PRC2 methylates histones and DNA / HDACs deacetylate histones / Ub-specific processing proteases / RNA Polymerase I Promoter Escape / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / Regulation of endogenous retroelements by KRAB-ZFP proteins / larval somatic muscle development / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / Senescence-Associated Secretory Phenotype (SASP) / Transcriptional regulation by small RNAs / Estrogen-dependent gene expression / HATs acetylate histones / UCH proteinases / Assembly of the ORC complex at the origin of replication / Oxidative Stress Induced Senescence / polytene chromosome / pre-miRNA export from nucleus / RNA nuclear export complex / snRNA import into nucleus / sulfate binding / manchette / mitotic nuclear membrane reassembly / cellular response to mineralocorticoid stimulus / Regulation of cholesterol biosynthesis by SREBP (SREBF) / importin-alpha family protein binding / Rev-mediated nuclear export of HIV RNA / Nuclear import of Rev protein / regulation of mitotic spindle assembly / protein localization to nucleolus / NEP/NS2 Interacts with the Cellular Export Machinery / tRNA processing in the nucleus / GTP metabolic process / Postmitotic nuclear pore complex (NPC) reformation / MicroRNA (miRNA) biogenesis / nucleosomal DNA binding / DNA metabolic process / regulation of mitotic nuclear division / nuclear chromosome / dynein intermediate chain binding / mitotic sister chromatid segregation / viral process / spermatid development / ribosomal large subunit export from nucleus / positive regulation of protein binding / sperm flagellum / nuclear pore / ribosomal subunit export from nucleus / nucleosome binding / spindle assembly / ribosomal small subunit export from nucleus / centriole / regulation of mitotic cell cycle / protein export from nucleus / guanyl-nucleotide exchange factor activity / condensed nuclear chromosome / mitotic spindle organization / male germ cell nucleus / hippocampus development / chromosome segregation / Transcriptional regulation by small RNAs / G1/S transition of mitotic cell cycle / recycling endosome / positive regulation of protein import into nucleus / small GTPase binding / protein import into nucleus / structural constituent of chromatin / GDP binding / melanosome / nuclear envelope / heterochromatin formation / nucleosome / mitotic cell cycle / nucleosome assembly / chromosome / G protein activity / chromatin organization / actin cytoskeleton organization / midbody / histone binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / cadherin binding / protein heterodimerization activity / protein domain specific binding / cell division / GTPase activity / chromatin binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
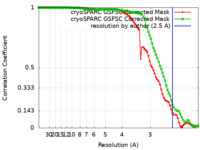
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Huang SK / Rubinstein JL / Kay LE | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Ran bound to RCC1 and the nucleosome core particle Authors: Huang SK / Rubinstein JL / Kay LE | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42685.map.gz emd_42685.map.gz | 14.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42685-v30.xml emd-42685-v30.xml emd-42685.xml emd-42685.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42685_fsc.xml emd_42685_fsc.xml emd_42685_fsc_2.xml emd_42685_fsc_2.xml | 8.4 KB 8.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_42685.png emd_42685.png | 64.8 KB | ||
| Filedesc metadata |  emd-42685.cif.gz emd-42685.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42685 http://ftp.pdbj.org/pub/emdb/structures/EMD-42685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42685 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42685 | HTTPS FTP |
-Validation report
| Summary document |  emd_42685_validation.pdf.gz emd_42685_validation.pdf.gz | 469.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42685_full_validation.pdf.gz emd_42685_full_validation.pdf.gz | 469.2 KB | Display | |
| Data in XML |  emd_42685_validation.xml.gz emd_42685_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_42685_validation.cif.gz emd_42685_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42685 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42685 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42685 | HTTPS FTP |
-Related structure data
| Related structure data |  8ux1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42685.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42685.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of the Ran-RCC1-NCP complex. The Ran-RCC1 portion was taken from a focus-refined Ran-RCC1 map. The NCP portion was taken from a 1-side bound Ran-RCC1-NCP map. Both local-sharpened. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Ran-RCC1-NCP complex
| Entire | Name: Ran-RCC1-NCP complex |
|---|---|
| Components |
|
-Supramolecule #1: Ran-RCC1-NCP complex
| Supramolecule | Name: Ran-RCC1-NCP complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.405036 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VMALQEASEA YLVGLFEDTN LSAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 Details: An extra isoleucine was inserted at the N-terminus following the first residue methionine Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.521611 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MITGRGKGGK GLGKGGAKRH RKVLRDNIQG ITKPAIRRLA RRGGVKRISG LIYEETRGVL KVFLENVIRD AVTYTEHAKR KTVTAMDVV YALKRQGRTL YGFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.388727 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKV KGKAKSRSNR AGLQFPVGRI HRLLRKGNYA ERVGAGAPVY LAAVMEYLAA EVLELAGNAA RDNKKTRIIP RHLQLAIRN DEELNKLLSG VTIAQGGVLP NIQAVLLPKK TEKKA UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 Details: An extra glycine was left at the N-terminus as a result of TEV cleavage. An extra isoleucine (residue 3) was inserted after the initiation methionine. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.897275 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GMIPPKTSGK AAKKAGKAQK NITKTDKKKK RKRKESYAIY IYKVLKQVHP DTGISSKAMS IMNSFVNDIF ERIAAEASRL AHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSSK UniProtKB: Histone H2B |
-Macromolecule #7: GTP-binding nuclear protein Ran
| Macromolecule | Name: GTP-binding nuclear protein Ran / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.456105 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAQGEPQVQ FKLVLVGDGG TGKTTFVKRH LTGEFEKKYV ATLGVEVHPL VFHTNRGPIK FNVWDTAGQE KFGGLRDGYY IQAQCAIIM FDVTSRVTYK NVPNWHRDLV RVCENIPIVL CGNKVDIKDR KVKAKSIVFH RKKNLQYYDI SAKSNYNFEK P FLWLARKL ...String: MAAQGEPQVQ FKLVLVGDGG TGKTTFVKRH LTGEFEKKYV ATLGVEVHPL VFHTNRGPIK FNVWDTAGQE KFGGLRDGYY IQAQCAIIM FDVTSRVTYK NVPNWHRDLV RVCENIPIVL CGNKVDIKDR KVKAKSIVFH RKKNLQYYDI SAKSNYNFEK P FLWLARKL IGDPNLEFVA MPALAPPEVV MDPALAAQYE HDLEVAQTTA LPDEDDDL UniProtKB: GTP-binding nuclear protein Ran |
-Macromolecule #8: Regulator of chromosome condensation
| Macromolecule | Name: Regulator of chromosome condensation / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.893758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPKRIAKRRS PPADAIPKSK KVKVSHRSHS TEPGLVLTLG QGDVGQLGLG ENVMERKKPA LVSIPEDVVQ AEAGGMHTVC LSKSGQVYS FGCNDEGALG RDTSVEGSEM VPGKVELQEK VVQVSAGDSH TAALTDDGRV FLWGSFRDNN GVIGLLEPMK K SMVPVQVQ ...String: SPKRIAKRRS PPADAIPKSK KVKVSHRSHS TEPGLVLTLG QGDVGQLGLG ENVMERKKPA LVSIPEDVVQ AEAGGMHTVC LSKSGQVYS FGCNDEGALG RDTSVEGSEM VPGKVELQEK VVQVSAGDSH TAALTDDGRV FLWGSFRDNN GVIGLLEPMK K SMVPVQVQ LDVPVVKVAS GNDHLVMLTA DGDLYTLGCG EQGQLGRVPE LFANRGGRQG LERLLVPKCV MLKSRGSRGH VR FQDAFCG AYFTFAISHE GHVYGFGLSN YHQLGTPGTE SCFIPQNLTS FKNSTKSWVG FSGGQHHTVC MDSEGKAYSL GRA EYGRLG LGEGAEEKSI PTLISRLPAV SSVACGASVG YAVTKDGRVF AWGMGTNYQL GTGQDEDAWS PVEMMGKQLE NRVV LSVSS GGQHTVLLVK DKEQS UniProtKB: Regulator of chromosome condensation |
-Macromolecule #5: 153-bp Widom 601 DNA forward strand
| Macromolecule | Name: 153-bp Widom 601 DNA forward strand / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 47.457234 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA) ...String: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DC)(DT) (DG)(DA)(DC)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG)(DC)(DG) (DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC)(DA)(DG) (DC)(DG)(DC) (DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DA)(DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT)(DC)(DG)(DG) (DC)(DA)(DC)(DC)(DG)(DG) (DG)(DA)(DT) (DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG)(DA)(DT) |
-Macromolecule #6: 153-bp Widom 601 DNA reverse strand
| Macromolecule | Name: 153-bp Widom 601 DNA reverse strand / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 46.998945 KDa |
| Sequence | String: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC)(DA) ...String: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC)(DA)(DG)(DC) (DT)(DC)(DT)(DA)(DG)(DC)(DA)(DC)(DC)(DG) (DC)(DT) (DT)(DA)(DA)(DA)(DC)(DG)(DC) (DA)(DC)(DG)(DT)(DA)(DC)(DG)(DC)(DG)(DC) (DT)(DG)(DT) (DC)(DC)(DC)(DC)(DC)(DG) (DC)(DG)(DT)(DT)(DT)(DT)(DA)(DA)(DC)(DC) (DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC)(DT) (DA)(DG)(DT)(DC)(DT) (DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC)(DG)(DT)(DG)(DT)(DC)(DA) (DG)(DA)(DT)(DA)(DT)(DA) (DT)(DA)(DC) (DA)(DT)(DC)(DC)(DT)(DG)(DT)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)