+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Site-one protease and SPRING | |||||||||
 Map data Map data | Primary map for SPRING-S1P | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | serine protease cholesterol metabolism zymogen activation Protein complex glycoprotein secretory pathway / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsite-1 protease / CREB3 factors activate genes / positive regulation of SREBP signaling pathway / ATF6-mediated unfolded protein response / regulation of cholesterol biosynthetic process / ATF6 (ATF6-alpha) activates chaperones / ATF6B (ATF6-beta) activates chaperones / Assembly of active LPL and LIPC lipase complexes / membrane protein intracellular domain proteolysis / regulation of vesicle-mediated transport ...site-1 protease / CREB3 factors activate genes / positive regulation of SREBP signaling pathway / ATF6-mediated unfolded protein response / regulation of cholesterol biosynthetic process / ATF6 (ATF6-alpha) activates chaperones / ATF6B (ATF6-beta) activates chaperones / Assembly of active LPL and LIPC lipase complexes / membrane protein intracellular domain proteolysis / regulation of vesicle-mediated transport / Regulation of cholesterol biosynthesis by SREBP (SREBF) / Golgi stack / lysosome organization / mitotic G2 DNA damage checkpoint signaling / endoplasmic reticulum unfolded protein response / cholesterol metabolic process / response to endoplasmic reticulum stress / protein maturation / Post-translational protein phosphorylation / protein processing / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / protein import into nucleus / endoplasmic reticulum lumen / Golgi membrane / serine-type endopeptidase activity / endoplasmic reticulum membrane / Golgi apparatus / proteolysis Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
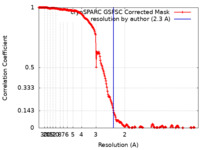
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Kober DL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: SPRING licenses S1P-mediated cleavage of SREBP2 by displacing an inhibitory pro-domain. Authors: Sebastian Hendrix / Vincent Dartigue / Hailee Hall / Shrankhla Bawaria / Jenina Kingma / Bilkish Bajaj / Noam Zelcer / Daniel L Kober /   Abstract: Site-one protease (S1P) conducts the first of two cleavage events in the Golgi to activate Sterol regulatory element binding proteins (SREBPs) and upregulate lipogenic transcription. S1P is also ...Site-one protease (S1P) conducts the first of two cleavage events in the Golgi to activate Sterol regulatory element binding proteins (SREBPs) and upregulate lipogenic transcription. S1P is also required for a wide array of additional signaling pathways. A zymogen serine protease, S1P matures through autoproteolysis of two pro-domains, with one cleavage event in the endoplasmic reticulum (ER) and the other in the Golgi. We recently identified the SREBP regulating gene, (SPRING), which enhances S1P maturation and is necessary for SREBP signaling. Here, we report the cryo-EM structures of S1P and S1P-SPRING at sub-2.5 Å resolution. SPRING activates S1P by dislodging its inhibitory pro-domain and stabilizing intra-domain contacts. Functionally, SPRING licenses S1P to cleave its cognate substrate, SREBP2. Our findings reveal an activation mechanism for S1P and provide insights into how spatial control of S1P activity underpins cholesterol homeostasis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42639.map.gz emd_42639.map.gz | 411.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42639-v30.xml emd-42639-v30.xml emd-42639.xml emd-42639.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42639_fsc.xml emd_42639_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_42639.png emd_42639.png | 127.4 KB | ||
| Filedesc metadata |  emd-42639.cif.gz emd-42639.cif.gz | 7.1 KB | ||
| Others |  emd_42639_additional_1.map.gz emd_42639_additional_1.map.gz emd_42639_half_map_1.map.gz emd_42639_half_map_1.map.gz emd_42639_half_map_2.map.gz emd_42639_half_map_2.map.gz | 422.2 MB 763.8 MB 763.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42639 http://ftp.pdbj.org/pub/emdb/structures/EMD-42639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42639 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42639 | HTTPS FTP |
-Validation report
| Summary document |  emd_42639_validation.pdf.gz emd_42639_validation.pdf.gz | 883.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42639_full_validation.pdf.gz emd_42639_full_validation.pdf.gz | 883.3 KB | Display | |
| Data in XML |  emd_42639_validation.xml.gz emd_42639_validation.xml.gz | 30 KB | Display | |
| Data in CIF |  emd_42639_validation.cif.gz emd_42639_validation.cif.gz | 39.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42639 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42639 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42639 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42639 | HTTPS FTP |
-Related structure data
| Related structure data |  8uw8MC  8uwcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42639.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42639.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map for SPRING-S1P | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.535 Å | ||||||||||||||||||||||||||||||||||||
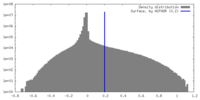
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharpened primary map, B factor = -70
| File | emd_42639_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened primary map, B factor = -70 | ||||||||||||
| Projections & Slices |
| ||||||||||||
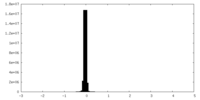
| Density Histograms |
-Half map: Half map A
| File | emd_42639_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
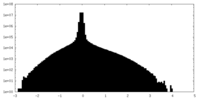
| Density Histograms |
-Half map: Half map B
| File | emd_42639_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
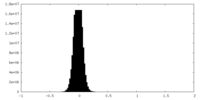
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of matured site-1 protease bound to SPRING
| Entire | Name: Complex of matured site-1 protease bound to SPRING |
|---|---|
| Components |
|
-Supramolecule #1: Complex of matured site-1 protease bound to SPRING
| Supramolecule | Name: Complex of matured site-1 protease bound to SPRING / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Co-expressed and secreted ectodomains of SPRING-His10 and S1P-FLAG purified using NiNTA chromatography and gel filtration. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Membrane-bound transcription factor site-1 protease
| Macromolecule | Name: Membrane-bound transcription factor site-1 protease / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 115.048203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKLVNIWLLL LVVLLCGKKH LGDRLEKKSF EKAPCPGCSH LTLKVEFSST VVEYEYIVAF NGYFTAKARN SFISSALKSS EVDNWRIIP RNNPSSDYPS DFEVIQIKEK QKAGLLTLED HPNIKRVTPQ RKVFRSLKYA ESDPTVPCNE TRWSQKWQSS R PLRRASLS ...String: MKLVNIWLLL LVVLLCGKKH LGDRLEKKSF EKAPCPGCSH LTLKVEFSST VVEYEYIVAF NGYFTAKARN SFISSALKSS EVDNWRIIP RNNPSSDYPS DFEVIQIKEK QKAGLLTLED HPNIKRVTPQ RKVFRSLKYA ESDPTVPCNE TRWSQKWQSS R PLRRASLS LGSGFWHATG RHSSRRLLRA IPRQVAQTLQ ADVLWQMGYT GANVRVAVFD TGLSEKHPHF KNVKERTNWT NE RTLDDGL GHGTFVAGVI ASMRECQGFA PDAELHIFRV FTNNQVSYTS WFLDAFNYAI LKKIDVLNLS IGGPDFMDHP FVD KVWELT ANNVIMVSAI GNDGPLYGTL NNPADQMDVI GVGGIDFEDN IARFSSRGMT TWELPGGYGR MKPDIVTYGA GVRG SGVKG GCRALSGTSV ASPVVAGAVT LLVSTVQKRE LVNPASMKQA LIASARRLPG VNMFEQGHGK LDLLRAYQIL NSYKP QASL SPSYIDLTEC PYMWPYCSQP IYYGGMPTVV NVTILNGMGV TGRIVDKPDW QPYLPQNGDN IEVAFSYSSV LWPWSG YLA ISISVTKKAA SWEGIAQGHV MITVASPAET ESKNGAEQTS TVKLPIKVKI IPTPPRSKRV LWDQYHNLRY PPGYFPR DN LRMKNDPLDW NGDHIHTNFR DMYQHLRSMG YFVEVLGAPF TCFDASQYGT LLMVDSEEEY FPEEIAKLRR DVDNGLSL V IFSDWYNTSV MRKVKFYDEN TRQWWMPDTG GANIPALNEL LSVWNMGFSD GLYEGEFTLA NHDMYYASGC SIAKFPEDG VVITQTFKDQ GLEVLKQETA VVENVPILGL YQIPAEGGGR IVLYGDSNCL DDSHRQKDCF WLLDALLQYT SYGVTPPSLS HSGNRQRPP SGAGSVTPER MEGNHLHRYS KVLEAHLGDP KPRPLPACPR LSWAKPQPLN ETAPSNLWKH QKLLSIDLDK V VLPNFRSN RPQVRPLSPG ESGAWDIPGG IMPGRYNQEV DYKDDDDKGS DYKDDDDKGS DYKDDDDK UniProtKB: Membrane-bound transcription factor site-1 protease |
-Macromolecule #2: SREBP regulating gene protein
| Macromolecule | Name: SREBP regulating gene protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.200291 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DKQEERAVRD RNLLQVHDHN QPIPWKVQFN LGNSSRPSNQ CRNSIQGKHL ITDELGYVCE RKDLLVNGC CNVNVPSTKQ YCCDGCWPNG CCSAYEYCVS CCLQPNKQLL LERFLNRAAV AFQNLFMAVE DHFELCLAKC R TSSQSVQH ...String: METDTLLLWV LLLWVPGSTG DKQEERAVRD RNLLQVHDHN QPIPWKVQFN LGNSSRPSNQ CRNSIQGKHL ITDELGYVCE RKDLLVNGC CNVNVPSTKQ YCCDGCWPNG CCSAYEYCVS CCLQPNKQLL LERFLNRAAV AFQNLFMAVE DHFELCLAKC R TSSQSVQH ENTYRDPIAK YCYGESPPEL FPAHHHHHHH HHH UniProtKB: SREBP regulating gene protein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 75 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 50 mM HEPES-NaOH and 150 mM NaCl | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa / Details: 30 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 4885 / Average exposure time: 7.0 sec. / Average electron dose: 48.0 e/Å2 / Details: CDS mode, 8 electrons per second, 50 frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)