+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of NaDC3-aKG complex | |||||||||||||||
 Map data Map data | Sharpened map of NaDC3-aKG in Ci-Ci conformation | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Na(+)/dicarboxylate cotransporter(NaDC3) / Solute carries / Elevator type alternating access / membrane protein / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationdicarboxylic acid transmembrane transporter activity / dicarboxylic acid transport / high-affinity sodium:dicarboxylate symporter activity / sodium:dicarboxylate symporter activity / citrate transmembrane transporter activity / citrate transport / succinate transmembrane transport / Sodium-coupled sulphate, di- and tri-carboxylate transporters / alpha-ketoglutarate transmembrane transporter activity / succinate transmembrane transporter activity ...dicarboxylic acid transmembrane transporter activity / dicarboxylic acid transport / high-affinity sodium:dicarboxylate symporter activity / sodium:dicarboxylate symporter activity / citrate transmembrane transporter activity / citrate transport / succinate transmembrane transport / Sodium-coupled sulphate, di- and tri-carboxylate transporters / alpha-ketoglutarate transmembrane transporter activity / succinate transmembrane transporter activity / glutathione transmembrane transport / glutathione transmembrane transporter activity / lipid transport / transport across blood-brain barrier / basolateral plasma membrane / extracellular exosome / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.09 Å | |||||||||||||||
 Authors Authors | Li Y / Wang DN / Mindell JA / Rice WJ / Song J / Mikusevic V / Marden JJ / Becerril A / Kuang H / Wang B | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Substrate translocation and inhibition in human dicarboxylate transporter NaDC3. Authors: Yan Li / Jinmei Song / Vedrana Mikusevic / Jennifer J Marden / Alissa Becerril / Huihui Kuang / Bing Wang / William J Rice / Joseph A Mindell / Da-Neng Wang /  Abstract: The human high-affinity sodium-dicarboxylate cotransporter (NaDC3) imports various substrates into the cell as tricarboxylate acid cycle intermediates, lipid biosynthesis precursors and signaling ...The human high-affinity sodium-dicarboxylate cotransporter (NaDC3) imports various substrates into the cell as tricarboxylate acid cycle intermediates, lipid biosynthesis precursors and signaling molecules. Understanding the cellular signaling process and developing inhibitors require knowledge of the structural basis of the dicarboxylate specificity and inhibition mechanism of NaDC3. To this end, we determined the cryo-electron microscopy structures of NaDC3 in various dimers, revealing the protomer in three conformations: outward-open C, outward-occluded C and inward-open C. A dicarboxylate is first bound and recognized in C and how the substrate interacts with NaDC3 in C likely helps to further determine the substrate specificity. A phenylalanine from the scaffold domain interacts with the bound dicarboxylate in the C state and modulates the kinetic barrier to the transport domain movement. Structural comparison of an inhibitor-bound structure of NaDC3 to that of the sodium-dependent citrate transporter suggests ways for making an inhibitor that is specific for NaDC3. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42615.map.gz emd_42615.map.gz | 87.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42615-v30.xml emd-42615-v30.xml emd-42615.xml emd-42615.xml | 29.3 KB 29.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42615_fsc.xml emd_42615_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42615.png emd_42615.png | 96.4 KB | ||
| Masks |  emd_42615_msk_1.map emd_42615_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42615.cif.gz emd-42615.cif.gz | 8.1 KB | ||
| Others |  emd_42615_additional_1.map.gz emd_42615_additional_1.map.gz emd_42615_half_map_1.map.gz emd_42615_half_map_1.map.gz emd_42615_half_map_2.map.gz emd_42615_half_map_2.map.gz | 55.5 MB 86.1 MB 86.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42615 http://ftp.pdbj.org/pub/emdb/structures/EMD-42615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42615 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42615 | HTTPS FTP |
-Validation report
| Summary document |  emd_42615_validation.pdf.gz emd_42615_validation.pdf.gz | 808.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42615_full_validation.pdf.gz emd_42615_full_validation.pdf.gz | 808.2 KB | Display | |
| Data in XML |  emd_42615_validation.xml.gz emd_42615_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_42615_validation.cif.gz emd_42615_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42615 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42615 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42615 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42615 | HTTPS FTP |
-Related structure data
| Related structure data |  8uvcMC  8uvbC  8uvdC  8uveC  8uvfC  8uvgC  8uvhC  8uviC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42615.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42615.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of NaDC3-aKG in Ci-Ci conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42615_msk_1.map emd_42615_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
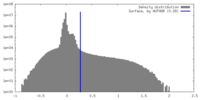
| Density Histograms |
-Additional map: Unsharpened map of NaDC3-aKG in Ci-Ci conformation
| File | emd_42615_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map of NaDC3-aKG in Ci-Ci conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
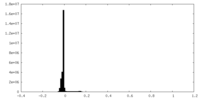
| Density Histograms |
-Half map: Half map A of NaDC3-aKG in Ci-Ci conformation
| File | emd_42615_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A of NaDC3-aKG in Ci-Ci conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B of NaDC3-aKG in Ci-Ci conformation
| File | emd_42615_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B of NaDC3-aKG in Ci-Ci conformation | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimer of NaDC3 in complex with alpha-ketoglutarate
| Entire | Name: Dimer of NaDC3 in complex with alpha-ketoglutarate |
|---|---|
| Components |
|
-Supramolecule #1: Dimer of NaDC3 in complex with alpha-ketoglutarate
| Supramolecule | Name: Dimer of NaDC3 in complex with alpha-ketoglutarate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.062 KDa |
-Macromolecule #1: Na(+)/dicarboxylate cotransporter 3
| Macromolecule | Name: Na(+)/dicarboxylate cotransporter 3 / type: protein_or_peptide / ID: 1 / Details: Solute carrier family 13 member 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.236023 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAALAAAAKK VWSARRLLVL LFTPLALLPV VFALPPKEGR CLFVILLMAV YWCTEALPLS VTALLPIVLF PFMGILPSNK VCPQYFLDT NFLFLSGLIM ASAIEEWNLH RRIALKILML VGVQPARLIL GMMVTTSFLS MWLSNTASTA MMLPIANAIL K SLFGQKEV ...String: MAALAAAAKK VWSARRLLVL LFTPLALLPV VFALPPKEGR CLFVILLMAV YWCTEALPLS VTALLPIVLF PFMGILPSNK VCPQYFLDT NFLFLSGLIM ASAIEEWNLH RRIALKILML VGVQPARLIL GMMVTTSFLS MWLSNTASTA MMLPIANAIL K SLFGQKEV RKDPSQESEE NTAAVRRNGL HTVPTEMQFL ASTEAKDHPG ETEVPLDLPA DSRKEDEYRR NIWKGFLISI PY SASIGGT ATLTGTAPNL ILLGQLKSFF PQCDVVNFGS WFIFAFPLML LFLLAGWLWI SFLYGGLSFR GWRKNKSEIR TNA EDRARA VIREEYQNLG PIKFAEQAVF ILFCMFAILL FTRDPKFIPG WASLFNPGFL SDAVTGVAIV TILFFFPSQR PSLK WWFDF KAPNTETEPL LTWKKAQETV PWNIILLLGG GFAMAKGCEE SGLSVWIGGQ LHPLENVPPA LAVLLITVVI AFFTE FASN TATIIIFLPV LAELAIRLRV HPLYLMIPGT VGCSFAFMLP VSTPPNSIAF ASGHLLVKDM VRTGLLMNLM GVLLLS LAM NTWAQTIFQL GTFPDWADMY SVNVTALPPT LANDTFRTLS GAGA UniProtKB: Na(+)/dicarboxylate cotransporter 3 |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: 2-OXOGLUTARIC ACID
| Macromolecule | Name: 2-OXOGLUTARIC ACID / type: ligand / ID: 3 / Number of copies: 2 / Formula: AKG |
|---|---|
| Molecular weight | Theoretical: 146.098 Da |
| Chemical component information |  ChemComp-AKG: |
-Macromolecule #4: TETRADECANE
| Macromolecule | Name: TETRADECANE / type: ligand / ID: 4 / Number of copies: 6 / Formula: C14 |
|---|---|
| Molecular weight | Theoretical: 198.388 Da |
| Chemical component information |  ChemComp-C14: |
-Macromolecule #5: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 5 / Number of copies: 4 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa / Details: Hold 10s before glow discharge | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 80.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.05 mrad |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 4201 / Average exposure time: 1.8 sec. / Average electron dose: 53.11 e/Å2 Details: 4201 untilted images were collected in super resolution mode at 40 frames per micrograph |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Chain ID: AB / Chain - Residue range: 1-602 / Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: The whole model was used as an initial model |
|---|---|
| Details | Initial local fitting was done using Chimera and then coot was used for ajustment. |
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 42.46 / Target criteria: cross-correlation coefficient |
| Output model |  PDB-8uvc: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)