[English] 日本語
 Yorodumi
Yorodumi- EMDB-42550: KIF1A[1-393] AMP-PNP bound one-head-bound state in complex with a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | KIF1A[1-393] AMP-PNP bound one-head-bound state in complex with a microtubule - class T1L02* | |||||||||||||||||||||
 Map data Map data | Primary map, locally refined on the two central asymmetric units | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | KIF1A / kinesin / motility / microtubule / tubulin / MOTOR PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationneuronal dense core vesicle membrane / plus-end-directed kinesin ATPase / dense core granule cytoskeletal transport / anterograde neuronal dense core vesicle transport / retrograde neuronal dense core vesicle transport / regulation of dendritic spine development / regulation of dendritic spine morphogenesis / anterograde axonal transport / plus-end-directed microtubule motor activity / Kinesins ...neuronal dense core vesicle membrane / plus-end-directed kinesin ATPase / dense core granule cytoskeletal transport / anterograde neuronal dense core vesicle transport / retrograde neuronal dense core vesicle transport / regulation of dendritic spine development / regulation of dendritic spine morphogenesis / anterograde axonal transport / plus-end-directed microtubule motor activity / Kinesins / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / kinesin complex / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / COPI-dependent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / cytoskeletal motor activity / neuronal dense core vesicle / microtubule-based process / vesicle-mediated transport / axon cytoplasm / structural constituent of cytoskeleton / microtubule cytoskeleton organization / mitotic cell cycle / microtubule cytoskeleton / microtubule binding / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / axon / GTPase activity / dendrite / synapse / GTP binding / perinuclear region of cytoplasm / ATP hydrolysis activity / ATP binding / metal ion binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Benoit MPMH / Rao L / Asenjo AB / Gennerich A / Sosa H | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L. Authors: Matthieu P M H Benoit / Lu Rao / Ana B Asenjo / Arne Gennerich / Hernando Sosa /  Abstract: Mutations in the microtubule-associated motor protein KIF1A lead to severe neurological conditions known as KIF1A-associated neurological disorders (KAND). Despite insights into its molecular ...Mutations in the microtubule-associated motor protein KIF1A lead to severe neurological conditions known as KIF1A-associated neurological disorders (KAND). Despite insights into its molecular mechanism, high-resolution structures of KIF1A-microtubule complexes remain undefined. Here, we present 2.7-3.5 Å resolution structures of dimeric microtubule-bound KIF1A, including the pathogenic P305L mutant, across various nucleotide states. Our structures reveal that KIF1A binds microtubules in one- and two-heads-bound configurations, with both heads exhibiting distinct conformations with tight inter-head connection. Notably, KIF1A's class-specific loop 12 (K-loop) forms electrostatic interactions with the C-terminal tails of both α- and β-tubulin. The P305L mutation does not disrupt these interactions but alters loop-12's conformation, impairing strong microtubule-binding. Structure-function analysis reveals the K-loop and head-head coordination as major determinants of KIF1A's superprocessive motility. Our findings advance the understanding of KIF1A's molecular mechanism and provide a basis for developing structure-guided therapeutics against KAND. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42550.map.gz emd_42550.map.gz | 33.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42550-v30.xml emd-42550-v30.xml emd-42550.xml emd-42550.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42550_fsc.xml emd_42550_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42550.png emd_42550.png | 121.3 KB | ||
| Masks |  emd_42550_msk_1.map emd_42550_msk_1.map emd_42550_msk_2.map emd_42550_msk_2.map emd_42550_msk_3.map emd_42550_msk_3.map emd_42550_msk_4.map emd_42550_msk_4.map | 274.6 MB 274.6 MB 274.6 MB 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42550.cif.gz emd-42550.cif.gz | 7.5 KB | ||
| Others |  emd_42550_additional_1.map.gz emd_42550_additional_1.map.gz emd_42550_additional_2.map.gz emd_42550_additional_2.map.gz emd_42550_half_map_1.map.gz emd_42550_half_map_1.map.gz emd_42550_half_map_2.map.gz emd_42550_half_map_2.map.gz | 236.1 MB 236.5 MB 219 MB 219 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42550 http://ftp.pdbj.org/pub/emdb/structures/EMD-42550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42550 | HTTPS FTP |
-Related structure data
| Related structure data |  8utuMC  8utnC  8utoC  8utpC  8utqC  8utrC  8utsC  8uttC  8utvC  8utwC  8utyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42550.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42550.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map, locally refined on the two central asymmetric units | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
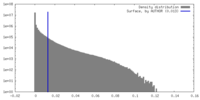
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42550_msk_1.map emd_42550_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_42550_msk_2.map emd_42550_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #3
| File |  emd_42550_msk_3.map emd_42550_msk_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #4
| File |  emd_42550_msk_4.map emd_42550_msk_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map low pass filtered to 8 A
| File | emd_42550_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map low pass filtered to 8 A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map low pass filtered to 6 A
| File | emd_42550_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map low pass filtered to 6 A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from the gold Standard refinement
| File | emd_42550_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from the gold Standard refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from the gold Standard refinement
| File | emd_42550_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from the gold Standard refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KIF1A[1-393] P305L mutant - AMP-PNP one and two-heads-bound state...
| Entire | Name: KIF1A[1-393] P305L mutant - AMP-PNP one and two-heads-bound states (merged) in complex with a 15R microtubule |
|---|---|
| Components |
|
-Supramolecule #1: KIF1A[1-393] P305L mutant - AMP-PNP one and two-heads-bound state...
| Supramolecule | Name: KIF1A[1-393] P305L mutant - AMP-PNP one and two-heads-bound states (merged) in complex with a 15R microtubule type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Macromolecule #1: Kinesin-like protein KIF1A
| Macromolecule | Name: Kinesin-like protein KIF1A / type: protein_or_peptide / ID: 1 Details: "linker" residues are comprised of a leucine zipper based on S. cerevisiae GCN4, which is followed by the C-terminal strep-tag Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.30448 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGASVKVAV RVRPFNSREM SRDSKCIIQM SGSTTTIVNP KQPKETPKSF SFDYSYWSHT SPEDINYASQ KQVYRDIGEE MLQHAFEGY NVCIFAYGQT GAGKSYTMMG KQEKDQQGII PQLCEDLFSR INDTTNDNMS YSVEVSYMEI YCERVRDLLN P KNKGNLRV ...String: MAGASVKVAV RVRPFNSREM SRDSKCIIQM SGSTTTIVNP KQPKETPKSF SFDYSYWSHT SPEDINYASQ KQVYRDIGEE MLQHAFEGY NVCIFAYGQT GAGKSYTMMG KQEKDQQGII PQLCEDLFSR INDTTNDNMS YSVEVSYMEI YCERVRDLLN P KNKGNLRV REHPLLGPYV EDLSKLAVTS YNDIQDLMDS GNKARTVAAT NMNETSSRSH AVFNIIFTQK RHDAETNITT EK VSKISLV DLAGSERADS TGAKGTRLKE GANINKSLTT LGKVISALAE MDSGPNKNKK KKKTDFILYR DSVLTWLLRE NLG GNSRTA MVAALSPADI NYDETLSTLR YADRAKQIRC NAVINEDPNN KLIRELKDEV TRLRDLLYAQ GLGDITDGAG VKQL EDKVE ELASKNYHLE NEVARLKKLV EFTSAWSHPQ FEK UniProtKB: Kinesin-like protein KIF1A |
-Macromolecule #2: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY UniProtKB: Tubulin alpha-1B chain |
-Macromolecule #3: Tubulin beta-2B chain
| Macromolecule | Name: Tubulin beta-2B chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.999887 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEATGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VMPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDSK NMMAACDPRH GRYLTVAAIF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATADEQGE FEEEEGEDEA UniProtKB: Tubulin beta chain |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #6: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #7: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #8: TAXOL
| Macromolecule | Name: TAXOL / type: ligand / ID: 8 / Number of copies: 1 / Formula: TA1 |
|---|---|
| Molecular weight | Theoretical: 853.906 Da |
| Chemical component information |  ChemComp-TA1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil / Support film - Material: GOLD | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.36 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.21 µm / Nominal defocus min: 0.81 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)