+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
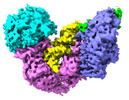
| Title | Spo11 core complex with gapped DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spo11 / Rec102 / Rec104 / Ski8 / DNA binding / Cross over. / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmeiotic DNA double-strand break processing / mRNA decay by 3' to 5' exoribonuclease / meiotic DNA double-strand break formation / protein-DNA complex assembly / Ski complex / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / nuclear-transcribed mRNA catabolic process, non-stop decay / DNA end binding / synaptonemal complex assembly / homologous chromosome pairing at meiosis ...meiotic DNA double-strand break processing / mRNA decay by 3' to 5' exoribonuclease / meiotic DNA double-strand break formation / protein-DNA complex assembly / Ski complex / nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay / nuclear-transcribed mRNA catabolic process, non-stop decay / DNA end binding / synaptonemal complex assembly / homologous chromosome pairing at meiosis / meiosis I / reciprocal meiotic recombination / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / nuclear chromosome / sporulation resulting in formation of a cellular spore / mRNA catabolic process / nuclear-transcribed mRNA catabolic process / condensed nuclear chromosome / site of double-strand break / protein-containing complex assembly / defense response to virus / chromatin binding / DNA binding / nucleoplasm / ATP binding / nucleus / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Yu Y / Patel DJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Cryo-EM structures of the Spo11 core complex bound to DNA. Authors: You Yu / Juncheng Wang / Kaixian Liu / Zhi Zheng / Meret Arter / Corentin Claeys Bouuaert / Stephen Pu / Dinshaw J Patel / Scott Keeney /    Abstract: DNA double-strand breaks that initiate meiotic recombination are formed by the topoisomerase-relative enzyme Spo11, supported by conserved auxiliary factors. Because high-resolution structural data ...DNA double-strand breaks that initiate meiotic recombination are formed by the topoisomerase-relative enzyme Spo11, supported by conserved auxiliary factors. Because high-resolution structural data have not been available, many questions remain about the architecture of Spo11 and its partners and how they engage with DNA. We report cryo-electron microscopy structures at up to 3.3-Å resolution of DNA-bound core complexes of Saccharomyces cerevisiae Spo11 with Rec102, Rec104 and Ski8. In these structures, monomeric core complexes make extensive contacts with the DNA backbone and with the recessed 3'-OH and first 5' overhanging nucleotide, establishing the molecular determinants of DNA end-binding specificity and providing insight into DNA cleavage preferences in vivo. The structures of individual subunits and their interfaces, supported by functional data in yeast, provide insight into the role of metal ions in DNA binding and uncover unexpected structural variation in homologs of the Top6BL component of the core complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42497.map.gz emd_42497.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42497-v30.xml emd-42497-v30.xml emd-42497.xml emd-42497.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42497.png emd_42497.png | 156.6 KB | ||
| Filedesc metadata |  emd-42497.cif.gz emd-42497.cif.gz | 6.9 KB | ||
| Others |  emd_42497_half_map_1.map.gz emd_42497_half_map_1.map.gz emd_42497_half_map_2.map.gz emd_42497_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42497 http://ftp.pdbj.org/pub/emdb/structures/EMD-42497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42497 | HTTPS FTP |
-Validation report
| Summary document |  emd_42497_validation.pdf.gz emd_42497_validation.pdf.gz | 774.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42497_full_validation.pdf.gz emd_42497_full_validation.pdf.gz | 774.3 KB | Display | |
| Data in XML |  emd_42497_validation.xml.gz emd_42497_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_42497_validation.cif.gz emd_42497_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42497 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42497 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42497 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42497 | HTTPS FTP |
-Related structure data
| Related structure data |  8urqMC  8uruC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42497.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42497.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map A
| File | emd_42497_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map B
| File | emd_42497_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Spo11 core complex bound to gapped DNA
| Entire | Name: Spo11 core complex bound to gapped DNA |
|---|---|
| Components |
|
-Supramolecule #1: Spo11 core complex bound to gapped DNA
| Supramolecule | Name: Spo11 core complex bound to gapped DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Meiotic recombination protein REC104
| Macromolecule | Name: Meiotic recombination protein REC104 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.763146 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIEEEDTNK ITCTQDFLHQ YFVTERVSIQ FGLNNKTVKR INKDEFDKAV NCIMSWTNYP KPGLKRTAST YLLSNSFKKS ATVSLPFIL GDPVCMPKRV ESNNNDTCLL YSDTLYDDPL IQRNDQAGDE IEDEFSFTLL RSEVNEIRPI SSSSTAQILQ S DYSALMYE RQASNGSIFQ FSSP UniProtKB: Meiotic recombination protein REC104 |
-Macromolecule #2: Meiotic recombination protein REC102
| Macromolecule | Name: Meiotic recombination protein REC102 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.263717 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARDITFLTV FLESCGAVNN DEAGKLLSAW TSTVRIEGPE STDSNSLYIP LLPPGMLKIK LNFKMNDRLV TEEQELFTKL REIVGSSIR FWEEQLFYQV QDVSTIENHV ILSLKCTILT DAQISTFISK PRELHTHAKG YPEIYYLSEL STTVNFFSKE G NYVEISQV ...String: MARDITFLTV FLESCGAVNN DEAGKLLSAW TSTVRIEGPE STDSNSLYIP LLPPGMLKIK LNFKMNDRLV TEEQELFTKL REIVGSSIR FWEEQLFYQV QDVSTIENHV ILSLKCTILT DAQISTFISK PRELHTHAKG YPEIYYLSEL STTVNFFSKE G NYVEISQV IPHFNEYFSS LIVSQLEFEY PMVFSMISRL RLKWQQSSLA PISYALTSNS VLLPIMLNMI AQDKSSTTAY QI LCRRRGP PIQNFQIFSL PAVTYNK UniProtKB: Meiotic recombination protein REC102 |
-Macromolecule #3: Meiosis-specific protein SPO11
| Macromolecule | Name: Meiosis-specific protein SPO11 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.020109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALEGLRKKY KTRQELVKAL TPKRRSIHLN SNGHSNGTPC SNADVLAHIK HFLSLAANSL EQHQQPISIV FQNKKKKGDT NSPDIHTTL DFPLNGPHLS THQFKLKRCA ILLNLLKVVM EKLPLGKNTT VRDIFYSNVE LFQRQANVVQ WLDVIRFNFK L SPRKSLNI ...String: MALEGLRKKY KTRQELVKAL TPKRRSIHLN SNGHSNGTPC SNADVLAHIK HFLSLAANSL EQHQQPISIV FQNKKKKGDT NSPDIHTTL DFPLNGPHLS THQFKLKRCA ILLNLLKVVM EKLPLGKNTT VRDIFYSNVE LFQRQANVVQ WLDVIRFNFK L SPRKSLNI IPAQKGLVYS PFPIDIYDNI LTCENEPKMQ KQTIFSGKPC LIPFFQDDAV IKLGTTSMCN IVIVEKEAVF TK LVNNYHK LSTNTMLITG KGFPDFLTRL FLKKLEQYCS NLISDCSIFT DADPYGISIA LNYTHSNERN AYICTMANYK GIR ITQVLA QNNEVHNKSI QLLSLNQRDY SLAKNLIASL TANSWDIATS PLKNVVIECQ REIFFQKKAE MNEIDAGIFK YKSR HHHHH HHHHHGDYKD DDDKDYKDDD DKDYKDDDDK UniProtKB: Meiosis-specific protein SPO11 |
-Macromolecule #4: Antiviral protein SKI8
| Macromolecule | Name: Antiviral protein SKI8 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.313555 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKVFIATAN AGKAHDADIF SVSACNSFTV SCSGDGYLKV WDNKLLDNEN PKDKSYSHFV HKSGLHHVDV LQTIERDAFE LCLVATTSF SGDLLFYRIT REDETKKVIF EKLDLLDSDM KKHSFWALKW GASNDRLLSH RLVATDVKGT TYIWKFHPFA D ESNSLTLN ...String: MSKVFIATAN AGKAHDADIF SVSACNSFTV SCSGDGYLKV WDNKLLDNEN PKDKSYSHFV HKSGLHHVDV LQTIERDAFE LCLVATTSF SGDLLFYRIT REDETKKVIF EKLDLLDSDM KKHSFWALKW GASNDRLLSH RLVATDVKGT TYIWKFHPFA D ESNSLTLN WSPTLELQGT VESPMTPSQF ATSVDISERG LIATGFNNGT VQISELSTLR PLYNFESQHS MINNSNSIRS VK FSPQGSL LAIAHDSNSF GCITLYETEF GERIGSLSVP THSSQASLGE FAHSSWVMSL SFNDSGETLC SAGWDGKLRF WDV KTKERI TTLNMHCDDI EIEEDILAVD EHGDSLAEPG VFDVKFLKKG WRSGMGADLN ESLCCVCLDR SIRWFREAGG K UniProtKB: Antiviral protein SKI8 |
-Macromolecule #5: gapped DNA
| Macromolecule | Name: gapped DNA / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.943064 KDa |
| Sequence | String: (DT)(DA)(DG)(DG)(DC)(DC)(DG)(DT)(DC)(DG) (DG)(DC)(DT)(DA)(DC)(DT)(DA)(DA)(DA)(DA) (DG)(DT)(DA)(DG)(DC)(DC)(DG)(DA)(DC) (DG)(DG)(DC)(DC)(DG)(DG)(DA)(DT)(DT)(DA) (DG) (DC)(DA)(DA)(DT)(DG)(DT) ...String: (DT)(DA)(DG)(DG)(DC)(DC)(DG)(DT)(DC)(DG) (DG)(DC)(DT)(DA)(DC)(DT)(DA)(DA)(DA)(DA) (DG)(DT)(DA)(DG)(DC)(DC)(DG)(DA)(DC) (DG)(DG)(DC)(DC)(DG)(DG)(DA)(DT)(DT)(DA) (DG) (DC)(DA)(DA)(DT)(DG)(DT)(DA)(DA) (DT)(DC)(DG)(DT)(DC)(DT)(DT)(DA)(DA)(DG) (DA)(DC) (DG)(DA)(DT)(DT)(DA)(DC)(DA) (DT)(DT)(DG)(DC) |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 25 mM HEPES, pH 7.4, 300 mM NaCl, 5 mM EDTA, 2 mM DTT |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 3.0 sec. / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 548674 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)