[English] 日本語
 Yorodumi
Yorodumi- EMDB-42429: Plasminogen binding group A streptococcus M-like protein from AP5... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasminogen binding group A streptococcus M-like protein from AP53 bound to human plasminogen | ||||||||||||
 Map data Map data | Full Map of hPg bound to PAM on a lentivirus | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | PAM / human plasminogen / M-protein / Group A streptococcus / BLOOD CLOTTING | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) | ||||||||||||
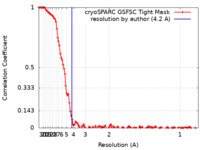
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | ||||||||||||
 Authors Authors | Readnour BM / Tjia-Fleck SK / Castellino FJ / McCann NR | ||||||||||||
| Funding support |  United States, 1 items United States, 1 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Plasminogen binding group A streptococcus M-like protein from AP53 bound to human plasminogen Authors: Readnour BM / Tjia-Fleck SK / McCann NR / Ayinuola YA / Ploplis VA / Castellino FJ | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42429.map.gz emd_42429.map.gz | 166.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42429-v30.xml emd-42429-v30.xml emd-42429.xml emd-42429.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42429_fsc.xml emd_42429_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_42429.png emd_42429.png | 80.7 KB | ||
| Filedesc metadata |  emd-42429.cif.gz emd-42429.cif.gz | 5 KB | ||
| Others |  emd_42429_half_map_1.map.gz emd_42429_half_map_1.map.gz emd_42429_half_map_2.map.gz emd_42429_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42429 http://ftp.pdbj.org/pub/emdb/structures/EMD-42429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42429 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42429 | HTTPS FTP |
-Related structure data
| Related structure data |  8tvlMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42429.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42429.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full Map of hPg bound to PAM on a lentivirus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.437 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map of hPg bound to PAM on a lentivirus
| File | emd_42429_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map of hPg bound to PAM on a lentivirus | ||||||||||||
| Projections & Slices |
| ||||||||||||
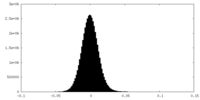
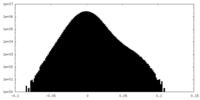
| Density Histograms |
-Half map: Half Map of hPg bound to PAM on a lentivirus
| File | emd_42429_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map of hPg bound to PAM on a lentivirus | ||||||||||||
| Projections & Slices |
| ||||||||||||
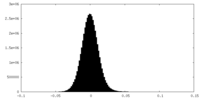
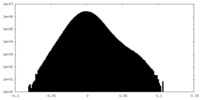
| Density Histograms |
- Sample components
Sample components
-Entire : Plasminogen Binding Group A Streptococcus M-Like Protein from AP5...
| Entire | Name: Plasminogen Binding Group A Streptococcus M-Like Protein from AP53 bound to human plasminogen |
|---|---|
| Components |
|
-Supramolecule #1: Plasminogen Binding Group A Streptococcus M-Like Protein from AP5...
| Supramolecule | Name: Plasminogen Binding Group A Streptococcus M-Like Protein from AP53 bound to human plasminogen type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: PAM was bound to the surface of a lentivirus |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) / Strain: AP53 / Location in cell: Cell surface Streptococcus pyogenes (bacteria) / Strain: AP53 / Location in cell: Cell surface |
| Molecular weight | Theoretical: 41 KDa |
-Macromolecule #1: Plasminogen binding group A streptococcus M-like protein
| Macromolecule | Name: Plasminogen binding group A streptococcus M-like protein type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) / Strain: AP53 Streptococcus pyogenes (bacteria) / Strain: AP53 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNRADDARNE VLRGNLVRAE LWYRQIQEND QL KLENKGL KTDLREKEEE LQGLKDDVEK LTADAELQRL KNERHEEAEL ERLKSERHDH DKK EAERKA LEDKLADKQE HLNGALRYIN EKEAEAKEKE AEQKKLKEEK QISDASRQGL RRDL DASRE AKKQVEKDLA ...String: MNRADDARNE VLRGNLVRAE LWYRQIQEND QL KLENKGL KTDLREKEEE LQGLKDDVEK LTADAELQRL KNERHEEAEL ERLKSERHDH DKK EAERKA LEDKLADKQE HLNGALRYIN EKEAEAKEKE AEQKKLKEEK QISDASRQGL RRDL DASRE AKKQVEKDLA NLTAELDKVK EEKQISDASR QGLRRDLDAS REAKKQVEKG LANLT AELD KVKEEKQISD ASRQGLRRDL DASREAKKQV EKALEEANSK LAALEKLNKE LEESKK LTE KEKAELQAKL EAEAKALKEQ LAKQAEELAK LRAEKASDSQ TPDAKPGNKA VPGKGQA PQ AGTKPNQNKA PMKETKRQLP STG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Formula: PBS |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | Plasminogen binding Group A streptococcus M like protein of AP53 bound to human plasminogen on the surface of a lentivirus |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 4054 / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 34.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 109000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8tvl: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)