+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4242 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Human cap-dependent 48S pre-initiation complex | |||||||||
 マップデータ マップデータ | human 48S pre-initiation complex, reconstituted in the presence of capped mRNA, eIF4B and eIF4F, displaying eIF2 ternary complex, eIF3 and eIF4B | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Translation initiation / 48S complex / capped mRNA / initiation factor 4B / start codon recognition / RIBOSOME | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報eukaryotic translation initiation factor 4F complex assembly / RNA strand-exchange activity / positive regulation of mRNA binding / viral translational termination-reinitiation / Cellular response to mitochondrial stress / eukaryotic translation initiation factor 3 complex, eIF3e / Response of EIF2AK1 (HRI) to heme deficiency / Recycling of eIF2:GDP / eukaryotic translation initiation factor 3 complex, eIF3m / Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S ...eukaryotic translation initiation factor 4F complex assembly / RNA strand-exchange activity / positive regulation of mRNA binding / viral translational termination-reinitiation / Cellular response to mitochondrial stress / eukaryotic translation initiation factor 3 complex, eIF3e / Response of EIF2AK1 (HRI) to heme deficiency / Recycling of eIF2:GDP / eukaryotic translation initiation factor 3 complex, eIF3m / Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S / methionyl-initiator methionine tRNA binding / RNA strand annealing activity / PERK regulates gene expression / eukaryotic translation initiation factor 2 complex / eukaryotic translation initiation factor 3 complex / eukaryotic translation initiation factor 4F complex / formation of cytoplasmic translation initiation complex / Z-decay: degradation of maternal mRNAs by zygotically expressed factors / cytoplasmic translational initiation / eukaryotic 43S preinitiation complex / translation factor activity, RNA binding / formation of translation preinitiation complex / Deadenylation of mRNA / eukaryotic 48S preinitiation complex / M-decay: degradation of maternal mRNAs by maternally stored factors / negative regulation of endoplasmic reticulum unfolded protein response / oxidized pyrimidine DNA binding / response to TNF agonist / positive regulation of base-excision repair / positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage / positive regulation of gastrulation / protein tyrosine kinase inhibitor activity / positive regulation of endodeoxyribonuclease activity / IRE1-RACK1-PP2A complex / positive regulation of Golgi to plasma membrane protein transport / TNFR1-mediated ceramide production / negative regulation of DNA repair / negative regulation of RNA splicing / supercoiled DNA binding / neural crest cell differentiation / protein-synthesizing GTPase / NF-kappaB complex / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / cysteine-type endopeptidase activator activity involved in apoptotic process / oxidized purine DNA binding / regulation of translational initiation / negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide / regulation of establishment of cell polarity / negative regulation of bicellular tight junction assembly / ubiquitin-like protein conjugating enzyme binding / negative regulation of phagocytosis / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / laminin receptor activity / negative regulation of ubiquitin protein ligase activity / protein kinase A binding / ion channel inhibitor activity / Ribosomal scanning and start codon recognition / pigmentation / Translation initiation complex formation / positive regulation of mitochondrial depolarization / positive regulation of T cell receptor signaling pathway / negative regulation of Wnt signaling pathway / positive regulation of activated T cell proliferation / negative regulation of translational frameshifting / TOR signaling / Protein hydroxylation / BH3 domain binding / SARS-CoV-1 modulates host translation machinery / regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / iron-sulfur cluster binding / regulation of cell division / cellular response to ethanol / mTORC1-mediated signalling / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / ribosomal small subunit binding / Eukaryotic Translation Termination / ubiquitin ligase inhibitor activity / positive regulation of GTPase activity / SRP-dependent cotranslational protein targeting to membrane / Response of EIF2AK4 (GCN2) to amino acid deficiency / protein serine/threonine kinase inhibitor activity / negative regulation of ubiquitin-dependent protein catabolic process / positive regulation of signal transduction by p53 class mediator / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / phagocytic cup / regulation of translational fidelity / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / negative regulation of protein binding / Nuclear events stimulated by ALK signaling in cancer / positive regulation of intrinsic apoptotic signaling pathway 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.3 Å | |||||||||
 データ登録者 データ登録者 | Schaffitzel C | |||||||||
| 資金援助 |  ベルギー, 2件 ベルギー, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nucleic Acids Res / 年: 2018 ジャーナル: Nucleic Acids Res / 年: 2018タイトル: Structure of a human cap-dependent 48S translation pre-initiation complex. 著者: Boris Eliseev / Lahari Yeramala / Alexander Leitner / Manikandan Karuppasamy / Etienne Raimondeau / Karine Huard / Elena Alkalaeva / Ruedi Aebersold / Christiane Schaffitzel /     要旨: Eukaryotic translation initiation is tightly regulated, requiring a set of conserved initiation factors (eIFs). Translation of a capped mRNA depends on the trimeric eIF4F complex and eIF4B to load ...Eukaryotic translation initiation is tightly regulated, requiring a set of conserved initiation factors (eIFs). Translation of a capped mRNA depends on the trimeric eIF4F complex and eIF4B to load the mRNA onto the 43S pre-initiation complex comprising 40S and initiation factors 1, 1A, 2, 3 and 5 as well as initiator-tRNA. Binding of the mRNA is followed by mRNA scanning in the 48S pre-initiation complex, until a start codon is recognised. Here, we use a reconstituted system to prepare human 48S complexes assembled on capped mRNA in the presence of eIF4B and eIF4F. The highly purified h-48S complexes are used for cross-linking/mass spectrometry, revealing the protein interaction network in this complex. We report the electron cryo-microscopy structure of the h-48S complex at 6.3 Å resolution. While the majority of eIF4B and eIF4F appear to be flexible with respect to the ribosome, additional density is detected at the entrance of the 40S mRNA channel which we attribute to the RNA-recognition motif of eIF4B. The eight core subunits of eIF3 are bound at the 40S solvent-exposed side, as well as the subunits eIF3d, eIF3b and eIF3i. elF2 and initiator-tRNA bound to the start codon are present at the 40S intersubunit side. This cryo-EM structure represents a molecular snap-shot revealing the h-48S complex following start codon recognition. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4242.map.gz emd_4242.map.gz | 32.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4242-v30.xml emd-4242-v30.xml emd-4242.xml emd-4242.xml | 74.1 KB 74.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_4242.png emd_4242.png | 56.4 KB | ||
| Filedesc metadata |  emd-4242.cif.gz emd-4242.cif.gz | 16.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4242 http://ftp.pdbj.org/pub/emdb/structures/EMD-4242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4242 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_4242_validation.pdf.gz emd_4242_validation.pdf.gz | 641.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_4242_full_validation.pdf.gz emd_4242_full_validation.pdf.gz | 641.1 KB | 表示 | |
| XML形式データ |  emd_4242_validation.xml.gz emd_4242_validation.xml.gz | 5 KB | 表示 | |
| CIF形式データ |  emd_4242_validation.cif.gz emd_4242_validation.cif.gz | 5.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4242 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4242 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4242.map.gz / 形式: CCP4 / 大きさ: 34.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4242.map.gz / 形式: CCP4 / 大きさ: 34.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | human 48S pre-initiation complex, reconstituted in the presence of capped mRNA, eIF4B and eIF4F, displaying eIF2 ternary complex, eIF3 and eIF4B | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
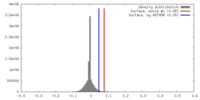
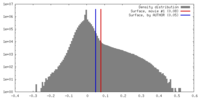
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : human cap-dependent 48S translation pre-initiation complex
+超分子 #1: human cap-dependent 48S translation pre-initiation complex
+超分子 #2: human cap-dependent 48S translation pre-initiation complex
+超分子 #3: mRNA
+超分子 #4: Eukaryotic translation initiation factor 4B
+分子 #1: Eukaryotic translation initiation factor 3 subunit A
+分子 #2: Eukaryotic translation initiation factor 3 subunit C
+分子 #3: Eukaryotic translation initiation factor 3 subunit E
+分子 #4: Eukaryotic translation initiation factor 3 subunit F
+分子 #5: Eukaryotic translation initiation factor 3 subunit H
+分子 #6: Eukaryotic translation initiation factor 3 subunit K
+分子 #7: Eukaryotic translation initiation factor 3 subunit L
+分子 #8: Eukaryotic translation initiation factor 3 subunit M
+分子 #9: EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT D
+分子 #12: 40S ribosomal protein S11
+分子 #13: 40S ribosomal protein S16
+分子 #14: 40S ribosomal protein S4, X isoform
+分子 #15: 40S ribosomal protein S29
+分子 #16: 40S ribosomal protein S9
+分子 #17: 40S ribosomal protein S18
+分子 #19: Eukaryotic translation initiation factor 2 subunit 1
+分子 #20: 40S ribosomal protein S23
+分子 #21: 40S ribosomal protein S19
+分子 #22: Eukaryotic translation initiation factor 2 subunit 3
+分子 #23: 40S ribosomal protein S5
+分子 #24: 40S ribosomal protein S30
+分子 #25: 40S ribosomal protein S25
+分子 #26: 40S ribosomal protein S7
+分子 #27: 40S ribosomal protein S27
+分子 #28: 40S ribosomal protein S13
+分子 #29: 40S ribosomal protein S15a
+分子 #30: 40S ribosomal protein S21
+分子 #31: 40S ribosomal protein S2
+分子 #32: EUKARYOTIC TRANSLATION INITIATION FACTOR 2 BETA SUBUNIT (eIF2-Beta)
+分子 #33: 40S ribosomal protein S17
+分子 #34: 40S ribosomal protein SA
+分子 #35: 40S ribosomal protein S3
+分子 #36: 40S ribosomal protein S20
+分子 #37: 40S ribosomal protein S3a
+分子 #38: 40S ribosomal protein S14
+分子 #39: 40S ribosomal protein S26
+分子 #40: 40S ribosomal protein S28
+分子 #41: Receptor of activated protein C kinase 1
+分子 #42: 40S ribosomal protein S15
+分子 #43: 40S ribosomal protein S8
+分子 #44: Ubiquitin-40S ribosomal protein S27a
+分子 #45: 40S ribosomal protein S6
+分子 #46: 40S ribosomal protein S12
+分子 #47: 40S ribosomal protein S24
+分子 #48: 40S ribosomal protein S10
+分子 #49: Eukaryotic translation initiation factor 4B
+分子 #50: Eukaryotic translation initiation factor 3 subunit B
+分子 #10: 18S ribosomal RNA
+分子 #11: Messenger RNA (26-MER)
+分子 #18: Transfer RNA (75-MER)
+分子 #51: water
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 詳細: 20 mM Tris HCl, 50 mM KOAc, 2.5 mM MgCl2, 2 mM DTT, 0.25 mM spermidine 0.25 mM GMPPNP |
|---|---|
| グリッド | モデル: Quantifoil R2/2 |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON II (4k x 4k) 検出モード: INTEGRATING / 平均電子線量: 30.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 4.0 µm / 最小 デフォーカス(公称値): 1.5 µm / 倍率(公称値): 112000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT 当てはまり具合の基準: Cross-correlation coefficient |
|---|---|
| 得られたモデル |  PDB-6fec: |
 ムービー
ムービー コントローラー
コントローラー











































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)