+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Helicobacter pylori dcagM PR | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | T4SS / protein translocation / MEMBRANE PROTEIN | ||||||||||||
| Biological species |  Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) | ||||||||||||
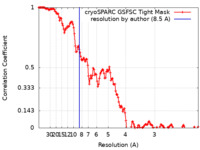
| Method | single particle reconstruction / cryo EM / Resolution: 8.5 Å | ||||||||||||
 Authors Authors | Roberts JR | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2024 Journal: Life Sci Alliance / Year: 2024Title: Subdomains of the Cag T4SS outer membrane core complex exhibit structural independence. Authors: Jacquelyn R Roberts / Sirena C Tran / Arwen E Frick-Cheng / Kaeli N Bryant / Chiamaka D Okoye / W Hayes McDonald / Timothy L Cover / Melanie D Ohi /  Abstract: The Cag type IV secretion system (Cag T4SS) has an important role in the pathogenesis of gastric cancer. The Cag T4SS outer membrane core complex (OMCC) is organized into three regions: a 14-fold ...The Cag type IV secretion system (Cag T4SS) has an important role in the pathogenesis of gastric cancer. The Cag T4SS outer membrane core complex (OMCC) is organized into three regions: a 14-fold symmetric outer membrane cap (OMC) composed of CagY, CagX, CagT, CagM, and Cag3; a 17-fold symmetric periplasmic ring (PR) composed of CagY and CagX; and a stalk with unknown composition. We investigated how CagT, CagM, and a conserved antenna projection (AP) region of CagY contribute to the structural organization of the OMCC. Single-particle cryo-EM analyses showed that complexes purified from Δ or Δ mutants no longer had organized OMCs, but the PRs remained structured. OMCCs purified from a CagY antenna projection mutant (CagYAP) were structurally similar to WT OMCCs, except for the absence of the α-helical antenna projection. These results indicate that CagY and CagX are sufficient for maintaining a stable PR, but the organization of the OMC requires CagY, CagX, CagM, and CagT. Our results highlight an unexpected structural independence of two major subdomains of the Cag T4SS OMCC. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42393.map.gz emd_42393.map.gz | 258.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42393-v30.xml emd-42393-v30.xml emd-42393.xml emd-42393.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42393_fsc.xml emd_42393_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42393.png emd_42393.png | 100.3 KB | ||
| Filedesc metadata |  emd-42393.cif.gz emd-42393.cif.gz | 5.9 KB | ||
| Others |  emd_42393_half_map_1.map.gz emd_42393_half_map_1.map.gz emd_42393_half_map_2.map.gz emd_42393_half_map_2.map.gz | 254.5 MB 254.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42393 http://ftp.pdbj.org/pub/emdb/structures/EMD-42393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42393 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42393.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42393.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42393_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
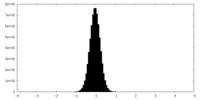
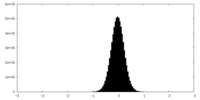
| Density Histograms |
-Half map: #1
| File | emd_42393_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
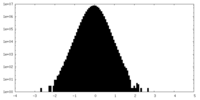
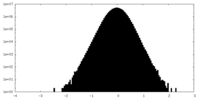
| Density Histograms |
- Sample components
Sample components
-Entire : Helicobacter pylori dcagM PR
| Entire | Name: Helicobacter pylori dcagM PR |
|---|---|
| Components |
|
-Supramolecule #1: Helicobacter pylori dcagM PR
| Supramolecule | Name: Helicobacter pylori dcagM PR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) / Strain: 26695 / Location in cell: membrane Helicobacter pylori 26695 (bacteria) / Strain: 26695 / Location in cell: membrane |
-Macromolecule #1: Cag pathogenicity island protein (Cag8)
| Macromolecule | Name: Cag pathogenicity island protein (Cag8) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) |
| Sequence | String: MGQAFFKKIV GCFCLGYLF L SSAIEAAA LD IKNFNRG RVK VVNKKI AYLG DEKPI TIWTS LDNV TVIQLE KDE TISYITT GF NKGWSIVP N SNHIFIQPK SVKSNLMFEK EAVNFALMT R DYQEFLKT KK LIVDAPD PKE LEEQKK ALEK EKEAK ...String: MGQAFFKKIV GCFCLGYLF L SSAIEAAA LD IKNFNRG RVK VVNKKI AYLG DEKPI TIWTS LDNV TVIQLE KDE TISYITT GF NKGWSIVP N SNHIFIQPK SVKSNLMFEK EAVNFALMT R DYQEFLKT KK LIVDAPD PKE LEEQKK ALEK EKEAK EQAQK AQKD KREKRK EER AKNRANL EN LTNAMSNP Q NLSNNKNLS EFIKQQRENE LDQMERLED M QEQAQANA LK QIEELNK KQA EETIKQ RAKD KINIK TDKPQ KSPE DNSIEL SPS DSAWRTN LV VRTNKALY Q FILRIAQKD NFASAYLTVK LEYPQRHEV S SVIEELKK RE EAKRQKE LIK QENLNT TAYI NRVMM ASNEQ IINK EKIREE KQK IILDQAK AL ETQYVHNA L KRNPVPRNY NYYQAPEKRS KHIMPSEIF D DGTFTYFG FK NITLQPA IFV VQPDGK LSMT DAAID PNMTN SGLR WYRVNE IAE KFKLIKD KA LVTVINKG Y GKNPLTKNY NIKNYGELER VIKKLPEVR D K |
-Macromolecule #2: Cag pathogenicity island protein (Cag7)
| Macromolecule | Name: Cag pathogenicity island protein (Cag7) / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) |
| Sequence | String: MNEENDKLET SKKAQQDSP Q DLSNEEAT EA NHFENLL KES KESSDH HLDN PTETQ THFDG DKSE ETQTQM DSE GNETSES SN GSLADKLF K KARKLVDNK KPFTQQKNLD EETQELNEE D DQENNEYQ EE TQTDLID DET SKKTQQ HSPQ DLSNE ...String: MNEENDKLET SKKAQQDSP Q DLSNEEAT EA NHFENLL KES KESSDH HLDN PTETQ THFDG DKSE ETQTQM DSE GNETSES SN GSLADKLF K KARKLVDNK KPFTQQKNLD EETQELNEE D DQENNEYQ EE TQTDLID DET SKKTQQ HSPQ DLSNE EATEA NHFE NLLKES KES SDHHLDN PT ETQTNFDG D KSEETQTQM DSEGNETSES SNGSLADKL F KKARKLVD NK KPFTQQK NLD EETQEL NEED DQENN EYQEE TQTD LIDDET SKK TQQHSPQ DL SNEEATEA N HFENLLKES KESSDHHLDN PTETQTNFD G DKSEEITD DS NDQEIIK GSK KKYIIG GIVV AVLIV IILFS RSIF HYFMPL EDK SSRFSKD RN LYVNDEIQ I RQEYNRLLK ERNEKGNMID KNLFFNDDP N RTLYNYLN IA EIEDKNP LRA FYECIS NGGN YEECL KLIKD KKLQ DQMKKT LEA YNDCIKN AK TEEERIKC L DLIKDENLK KSLLNQQKVQ VALDCLKNA K TDEERNEC LK LINDPEI REK FRKELE LQKE LQEYK DCIKN AKTE AEKNKC LKG LSKEAIE RL KQQALDCL K NAKTDEERN ECLKNIPQDL QKELLADMS V KAYKDCVS KA RNEKEKQ ECE KLLTPE ARKK LEQQV LDCLK NAKT DEERKK CLK DLPKDLQ SD ILAKESLK A YKDCVSQAK TEAEKKECEK LLTPEAKKL L EEEAKESV KA YLDCVSQ AKT EAEKKE CEKL LTPEA KKKLE EAKK SVKAYL DCV SRARNEK EK KECEKLLT P EAKKLLEQQ ALDCLKNAKT DKERKKCLK D LPKDLQKK VL AKESVKA YLD CVSQAK TEAE KKECE KLLTP EARK LLEEAK KSV KAYLDCV SQ AKTEAEKK E CEKLLTPEA RKLLEEXAKE SVKAYLDCV S QAKNEAEK KE CEKLLTL ESK KKLEEA KKSV KAYLD CVSQA KTEA EKKECE KLL TPEAKKL LE QQALDCLK N AKTEADKKR CVKDLPKDLQ KKVLAKESL K AYKDCVSK AR NEKEKKE CEK LLTPEA KKLL EEAKK SVKAY LDCV SQAKTE AEK KECEKLL TP EARKLLEE A KESVKAYKD CVSKARNEKE KKECEKLLT P EAKKLLEQ QV LDCLKNA KTE ADKKRC VKDL PKDLQ KKVLA KESV KAYLDC VSR ARNEKEK KE CEKLLTPE A KKLLEEAKE SLKAYKDCLS QARNEEERR A CEKLLTPE AR KLLEQEV KKS IKAYLD CVSR ARNEK EKKEC EKLL TPEARK FLA KQVLNCL EK AGNEEERK A CLKNLPKDL QENILAKESL KAYKDCLSQ A RNEEERRA CE KLLTPEA RKL LEQEVK KSVK AYLDC VSRAR NEKE KKECEK LLT PEARKFL AK ELQQKDKA I KDCLKNADP NDRAAIMKCL DGLSDEEKL K YLQEAREK AV ADCLAMA KTD EEKRKC QNLY SDLIQ EIQNK RTQN KQNQLS KTE RLHQASE CL DNLDDPTD Q EAIEQCLEG LSDSERALIL GIKRQADEV D LIYSDLRN RK TFDNMAA KGY PLLPMD FKNG GDIAT INATN VDAD KIASDN PIY ASIEPDI AK QYETEKTI K DKNLEAKLA KALGGNKKDD DKEKSKKST A EAKAENNK ID KDVAETA KNI SEIALK NKKE KSGEF VDENG NPID DKKKAE KQD ETSPVKQ AF IGKSDPTF V LAQYTPIEI TLTSKVDATL TGIVSGVVA K DVWNMNGT MI LLDKGTK VYG NYQSVK GGTP IMTRL MIVFT KAIT PDGVII PLA NAQAAGM LG EAGVDGYV N NHFMKRIGF AVIASVVNSF LQTAPIIAL D KLIGLGKG RS ERTPEFN YAL GQAING SMQS SAQMS NQILG QLMN IPPSFY KNE GDSIKIL TM DDIDFSGV Y DVKITNKSV VDEIIKQSTK TLSREHEEI T TSPKGGN |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)