[English] 日本語
 Yorodumi
Yorodumi- EMDB-42375: Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), cryo-EM -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
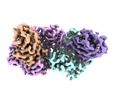
| Title | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), cryo-EM | ||||||||||||||||||
 Map data Map data | Refinement map. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | bacterial C-glucosyl deglycosidase / C-C bond cleavage / C-glucosyl aromatic polyketides / C-glucosyl flavonoids / Deinococcus aerius / soil bacterium / N-terminal DUF6379 beta-sandwich domain / C-terminal TIM-barrel domain / alpha2beta2 heterotetramer / LYASE | ||||||||||||||||||
| Function / homology | Domain of unknown function DUF6379 / C-glycoside deglycosidase beta subunit / : / Xylose isomerase-like, TIM barrel domain / Xylose isomerase-like TIM barrel / Xylose isomerase-like superfamily / lyase activity / Xylose isomerase-like TIM barrel domain-containing protein / C-deglycosylation enzyme beta subunit Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  Deinococcus aerius (bacteria) Deinococcus aerius (bacteria) | ||||||||||||||||||
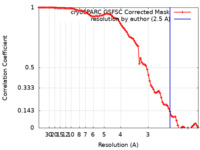
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||||||||||||||
 Authors Authors | Furlanetto V / Kalyani DC / Kostelac A / Haltrich D / Hallberg BM / Divne C | ||||||||||||||||||
| Funding support |  Sweden, Sweden,  Austria, 5 items Austria, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius. Authors: Valentina Furlanetto / Dayanand C Kalyani / Anja Kostelac / Jolanta Puc / Dietmar Haltrich / B Martin Hällberg / Christina Divne /   Abstract: Plant C-glycosylated aromatic polyketides are important for plant and animal health. These are specialized metabolites that perform functions both within the plant, and in interaction with soil or ...Plant C-glycosylated aromatic polyketides are important for plant and animal health. These are specialized metabolites that perform functions both within the plant, and in interaction with soil or intestinal microbes. Despite the importance of these plant compounds, there is still limited knowledge of how they are metabolized. The Gram-positive aerobic soil bacterium Deinococcus aerius strain TR0125 and other Deinococcus species thrive in a wide range of harsh environments. In this work, we identified a C-glycoside deglycosylation gene cluster in the genome of D. aerius. The cluster includes three genes coding for a GMC-type oxidoreductase (DaCGO1) that oxidizes the glucosyl C3 position in aromatic C-glucosyl compounds, which in turn provides the substrate for the C-glycoside deglycosidase (DaCGD; composed of α+β subunits) that cleaves the glucosyl-aglycone C-C bond. Our results from size-exclusion chromatography, single particle cryo-electron microscopy and X-ray crystallography show that DaCGD is an αβ heterotetramer, which represents a novel oligomeric state among bacterial CGDs. Importantly, the high-resolution X-ray structure of DaCGD provides valuable insights into the activation of the catalytic hydroxide ion by Lys261. DaCGO1 is specific for the 6-C-glucosyl flavones isovitexin, isoorientin and the 2-C-glucosyl xanthonoid mangiferin, and the subsequent C-C-bond cleavage by DaCGD generated apigenin, luteolin and norathyriol, respectively. Of the substrates tested, isovitexin was the preferred substrate (DaCGO1, K 0.047 mM, k 51 min; DaCGO1/DaCGD, K 0.083 mM, k 0.42 min). | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42375.map.gz emd_42375.map.gz | 55.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42375-v30.xml emd-42375-v30.xml emd-42375.xml emd-42375.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42375_fsc.xml emd_42375_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_42375.png emd_42375.png | 96.2 KB | ||
| Masks |  emd_42375_msk_1.map emd_42375_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42375.cif.gz emd-42375.cif.gz | 6.4 KB | ||
| Others |  emd_42375_half_map_1.map.gz emd_42375_half_map_1.map.gz emd_42375_half_map_2.map.gz emd_42375_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42375 http://ftp.pdbj.org/pub/emdb/structures/EMD-42375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42375 | HTTPS FTP |
-Related structure data
| Related structure data |  8umcMC  8qvcC  8qvdC  8qveC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42375.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42375.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||
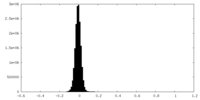
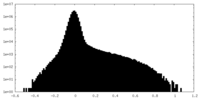
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42375_msk_1.map emd_42375_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap B
| File | emd_42375_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap A
| File | emd_42375_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C-glucosyl deglycosidase, alpha subunit
| Entire | Name: C-glucosyl deglycosidase, alpha subunit |
|---|---|
| Components |
|
-Supramolecule #1: C-glucosyl deglycosidase, alpha subunit
| Supramolecule | Name: C-glucosyl deglycosidase, alpha subunit / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The complex contains two alpha subunits and two beta subunits that form an alpha2beta2 heteroteramer |
|---|---|
| Source (natural) | Organism:  Deinococcus aerius (bacteria) / Strain: TR0125 Deinococcus aerius (bacteria) / Strain: TR0125 |
-Macromolecule #1: Xylose isomerase-like TIM barrel domain-containing protein
| Macromolecule | Name: Xylose isomerase-like TIM barrel domain-containing protein type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus aerius (bacteria) / Strain: TR0125 Deinococcus aerius (bacteria) / Strain: TR0125 |
| Molecular weight | Theoretical: 40.554328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQPQIKRGV SLYSFQEEFF LRKMTLEDCV AACASMGAYG IESLAEQMMP GFPNLDDAFY DGWHAMMAKY GTVSVCHDMF LDTKKFRGR LMTLDEQVES FVRDIRHASR LGCTVIRVLN FVSPELMEKV LPHAEQSNMR LGLEIHAPMH FEHPWVLRHI E FMDRLGSP ...String: MTQPQIKRGV SLYSFQEEFF LRKMTLEDCV AACASMGAYG IESLAEQMMP GFPNLDDAFY DGWHAMMAKY GTVSVCHDMF LDTKKFRGR LMTLDEQVES FVRDIRHASR LGCTVIRVLN FVSPELMEKV LPHAEQSNMR LGLEIHAPMH FEHPWVLRHI E FMDRLGSP LLGFIPDMGI FTKHFPPVMA ERLIRQGATP HIIEYIREQY DRRVLAEYVV GDVRNMGGNP VDIRAAEMLR HN NWSNPRR LLEHMDRIFH VHAKFYEMDE QDRETSLGYE EVIPVLKEGG YSGYLASEYE GNRHIQDAFE VDSVEQVRRH QRM LARLIG EREVAHVAEN LYFQSHHHHH H UniProtKB: Xylose isomerase-like TIM barrel domain-containing protein |
-Macromolecule #2: DUF6379 domain-containing protein
| Macromolecule | Name: DUF6379 domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Deinococcus aerius (bacteria) / Strain: TR0125 Deinococcus aerius (bacteria) / Strain: TR0125 |
| Molecular weight | Theoretical: 13.755647 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFDKYIVVED SLKRVPGGVQ FGVRLPYYRG LGLSMVETMD VTVDGERVPE ENLTVTLGDR TVPFARRDDE TDTIWNFGEI ATVTARLPH ELGPGEHQVG VNFGLRISYF PVPMVGQDAK TLKL UniProtKB: C-deglycosylation enzyme beta subunit |
-Macromolecule #3: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Details | Data collected at the Karolinska Institutet 3D-EM facility |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 2153 / Average exposure time: 1.5 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 20.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)