[English] 日本語
 Yorodumi
Yorodumi- EMDB-42268: Cryo-EM Structure of the Ro5256390-bound hTA1-Gs heterotrimer sig... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Ro5256390-bound hTA1-Gs heterotrimer signaling complex | |||||||||||||||
 Map data Map data | Primary Map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | GPCR / Signaling Complex / Trace Amine-associated Receptor / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCOPI-coated Golgi to ER transport vesicle / regulation of parathyroid hormone secretion / post-embryonic body morphogenesis / Amine ligand-binding receptors / adenylate cyclase-activating serotonin receptor signaling pathway / response to parathyroid hormone / trace-amine receptor activity / sensory perception of chemical stimulus / endochondral ossification / positive regulation of sodium ion transport ...COPI-coated Golgi to ER transport vesicle / regulation of parathyroid hormone secretion / post-embryonic body morphogenesis / Amine ligand-binding receptors / adenylate cyclase-activating serotonin receptor signaling pathway / response to parathyroid hormone / trace-amine receptor activity / sensory perception of chemical stimulus / endochondral ossification / positive regulation of sodium ion transport / tissue homeostasis / energy reserve metabolic process / genomic imprinting / embryonic cranial skeleton morphogenesis / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / positive regulation of osteoclast differentiation / positive regulation of mini excitatory postsynaptic potential / beta2-adrenergic receptor activity / negative regulation of smooth muscle contraction / AMPA selective glutamate receptor signaling pathway / cartilage development / norepinephrine-epinephrine-mediated vasodilation involved in regulation of systemic arterial blood pressure / positive regulation of autophagosome maturation / heat generation / norepinephrine binding / embryonic hindlimb morphogenesis / Adrenoceptors / beta-2 adrenergic receptor binding / positive regulation of lipophagy / negative regulation of G protein-coupled receptor signaling pathway / negative regulation of multicellular organism growth / response to psychosocial stress / adrenergic receptor signaling pathway / endosome to lysosome transport / skin development / diet induced thermogenesis / alkylglycerophosphoethanolamine phosphodiesterase activity / positive regulation of cAMP/PKA signal transduction / adenylate cyclase binding / smooth muscle contraction / developmental growth / hair follicle placode formation / G-protein alpha-subunit binding / regulation of signal transduction / bone resorption / potassium channel regulator activity / positive regulation of bone mineralization / positive regulation of osteoblast differentiation / D1 dopamine receptor binding / neuronal dense core vesicle / alpha-tubulin binding / intercellular bridge / negative regulation of blood pressure / regulation of sodium ion transport / adenylate cyclase-activating adrenergic receptor signaling pathway / ruffle / insulin-like growth factor receptor binding / response to prostaglandin E / endomembrane system / ionotropic glutamate receptor binding / adenylate cyclase regulator activity / brown fat cell differentiation / cellular response to glucagon stimulus / receptor-mediated endocytosis / response to cold / adenylate cyclase activator activity / trans-Golgi network membrane / post-embryonic development / skeletal system development / clathrin-coated endocytic vesicle membrane / electron transport chain / positive regulation of insulin secretion / recycling endosome / sarcolemma / bone development / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / multicellular organism growth / cognition / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / cellular response to amyloid-beta / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||||||||
 Authors Authors | Zilberg G / Warren AL / Parpounas AK / Wacker D | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular basis of human trace amine-associated receptor 1 activation. Authors: Gregory Zilberg / Alexandra K Parpounas / Audrey L Warren / Shifan Yang / Daniel Wacker /  Abstract: The human trace amine-associated receptor 1 (hTAAR1, hTA1) is a key regulator of monoaminergic neurotransmission and the actions of psychostimulants. Despite preclinical research demonstrating its ...The human trace amine-associated receptor 1 (hTAAR1, hTA1) is a key regulator of monoaminergic neurotransmission and the actions of psychostimulants. Despite preclinical research demonstrating its tractability as a drug target, its molecular mechanisms of activation remain unclear. Moreover, poorly understood pharmacological differences between rodent and human TA1 complicate the translation of findings from preclinical disease models into novel pharmacotherapies. To elucidate hTA1's mechanisms on the molecular scale and investigate the underpinnings of its divergent pharmacology from rodent orthologs, we herein report the structure of the human TA1 receptor in complex with a Gαs heterotrimer. Our structure reveals shared structural elements with other TAARs, as well as with its closest monoaminergic orthologue, the serotonin receptor 5-HT4R. We further find that a single mutation dramatically shifts the selectivity of hTA1 towards that of its rodent orthologues, and report on the effects of substituting residues to those found in serotonin and dopamine receptors. Strikingly, we also discover that the atypical antipsychotic medication and pan-monoaminergic antagonist asenapine potently and efficaciously activates hTA1. Together our studies provide detailed insight into hTA1 structure and function, contrast its molecular pharmacology with that of related receptors, and uncover off-target activities of monoaminergic drugs at hTA1. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42268.map.gz emd_42268.map.gz | 31.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42268-v30.xml emd-42268-v30.xml emd-42268.xml emd-42268.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42268.png emd_42268.png | 44.3 KB | ||
| Filedesc metadata |  emd-42268.cif.gz emd-42268.cif.gz | 7 KB | ||
| Others |  emd_42268_half_map_1.map.gz emd_42268_half_map_1.map.gz emd_42268_half_map_2.map.gz emd_42268_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42268 http://ftp.pdbj.org/pub/emdb/structures/EMD-42268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42268 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42268 | HTTPS FTP |
-Related structure data
| Related structure data |  8uhbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42268.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42268.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.069 Å | ||||||||||||||||||||||||||||||||||||
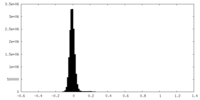
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 1
| File | emd_42268_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
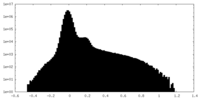
| Density Histograms |
-Half map: Half map 2
| File | emd_42268_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hTA1-Gs-Nb35 complex (Ro5256390)
| Entire | Name: hTA1-Gs-Nb35 complex (Ro5256390) |
|---|---|
| Components |
|
-Supramolecule #1: hTA1-Gs-Nb35 complex (Ro5256390)
| Supramolecule | Name: hTA1-Gs-Nb35 complex (Ro5256390) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Soluble cytochrome b562,Beta-2 adrenergic receptor,Trace amine-as...
| Macromolecule | Name: Soluble cytochrome b562,Beta-2 adrenergic receptor,Trace amine-associated receptor 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.019816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTTA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLMGQP G NGSAFLLA ...String: MKTIIALSYI FCLVFADYKD DDDAKLQTMH HHHHHHHHHE NLYFQGGTTA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLMGQP G NGSAFLLA PNRSHAPDHD VTQQRDEMMP FCHNIINISC VKNNWSNDVR ASLYSLMVLI ILTTLVGNLI VIVSISHFKQ LH TPTNWLI HSMATVDFLL GCLVMPYSMV RSAEHCWYFG EVFCKIHTST DIMLSSASIW HLSFISIDRY YAVCDPLRYK AKM NILVIC VMIFISWSVP AVFAFGMIFL ELNFKGAEEI YYKHVHCRGG CSVFFSKISG VLTFMTSFYI PGSIMLCVYY RIYL IAKEQ ARLISDANQK LQIGLEMKNG ISQSKERKAV KTLGIVMGVF LICWCPFFIC TVMDPFLHYI IPPTLNDVLI WFGYL NSTF NPMVYAFFYP WFRKALKMML FGKIFQKDSS RCKLFLELSS UniProtKB: Soluble cytochrome b562, Beta-2 adrenergic receptor, Trace amine-associated receptor 1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.803598 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPN FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKSTIVKQM RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPN FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA EYMPTEQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ EALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCS VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.71432 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSSH HHHHH |
-Macromolecule #6: (2R,4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-amine
| Macromolecule | Name: (2R,4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-amine / type: ligand / ID: 6 / Number of copies: 1 / Formula: WV8 |
|---|---|
| Molecular weight | Theoretical: 218.295 Da |
| Chemical component information |  ChemComp-WV8: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 53.88 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.35 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 626370 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


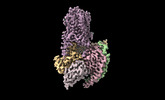

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)