[English] 日本語
 Yorodumi
Yorodumi- EMDB-42220: Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of bovine phosphodiesterase 6 bound to udenafil | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phosphodiesterase / GPCR effector enzyme / SIGNALING PROTEIN / inhibitor / SIGNALING PROTEIN-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology information3',5'-cyclic-GMP phosphodiesterase / Inactivation, recovery and regulation of the phototransduction cascade / Activation of the phototransduction cascade / positive regulation of G protein-coupled receptor signaling pathway / positive regulation of epidermal growth factor receptor signaling pathway / Ca2+ pathway / photoreceptor outer segment membrane / entrainment of circadian clock by photoperiod / cGMP binding / 3',5'-cyclic-GMP phosphodiesterase activity ...3',5'-cyclic-GMP phosphodiesterase / Inactivation, recovery and regulation of the phototransduction cascade / Activation of the phototransduction cascade / positive regulation of G protein-coupled receptor signaling pathway / positive regulation of epidermal growth factor receptor signaling pathway / Ca2+ pathway / photoreceptor outer segment membrane / entrainment of circadian clock by photoperiod / cGMP binding / 3',5'-cyclic-GMP phosphodiesterase activity / 3',5'-cyclic-AMP phosphodiesterase activity / : / visual perception / photoreceptor disc membrane / retina development in camera-type eye / molecular adaptor activity / positive regulation of MAPK cascade / zinc ion binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
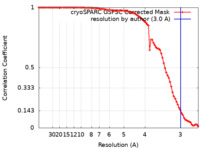
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Aplin C / Cerione RA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: Probing the mechanism by which the retinal G protein transducin activates its biological effector PDE6. Authors: Cody Aplin / Richard A Cerione /  Abstract: Phototransduction in retinal rods occurs when the G protein-coupled photoreceptor rhodopsin triggers the activation of phosphodiesterase 6 (PDE6) by GTP-bound alpha subunits of the G protein ...Phototransduction in retinal rods occurs when the G protein-coupled photoreceptor rhodopsin triggers the activation of phosphodiesterase 6 (PDE6) by GTP-bound alpha subunits of the G protein transducin (Gα). Recently, we presented a cryo-EM structure for a complex between two GTP-bound recombinant Gα subunits and native PDE6, that included a bivalent antibody bound to the C-terminal ends of Gα and the inhibitor vardenafil occupying the active sites on the PDEα and PDEβ subunits. We proposed Gα-activated PDE6 by inducing a striking reorientation of the PDEγ subunits away from the catalytic sites. However, questions remained including whether in the absence of the antibody Gα binds to PDE6 in a similar manner as observed when the antibody is present, does Gα activate PDE6 by enabling the substrate cGMP to access the catalytic sites, and how does the lipid membrane enhance PDE6 activation? Here, we demonstrate that 2:1 Gα-PDE6 complexes form with either recombinant or retinal Gα in the absence of the Gα antibody. We show that Gα binding is not necessary for cGMP nor competitive inhibitors to access the active sites; instead, occupancy of the substrate binding sites enables Gα to bind and reposition the PDE6γ subunits to promote catalytic activity. Moreover, we demonstrate by reconstituting Gα-stimulated PDE6 activity in lipid bilayer nanodiscs that the membrane-induced enhancement results from an increase in the apparent binding affinity of Gα for PDE6. These findings provide new insights into how the retinal G protein stimulates rapid catalytic turnover by PDE6 required for dim light vision. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42220.map.gz emd_42220.map.gz | 32.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42220-v30.xml emd-42220-v30.xml emd-42220.xml emd-42220.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42220_fsc.xml emd_42220_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_42220.png emd_42220.png | 109.8 KB | ||
| Filedesc metadata |  emd-42220.cif.gz emd-42220.cif.gz | 6.4 KB | ||
| Others |  emd_42220_half_map_1.map.gz emd_42220_half_map_1.map.gz emd_42220_half_map_2.map.gz emd_42220_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42220 http://ftp.pdbj.org/pub/emdb/structures/EMD-42220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42220 | HTTPS FTP |
-Validation report
| Summary document |  emd_42220_validation.pdf.gz emd_42220_validation.pdf.gz | 697 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42220_full_validation.pdf.gz emd_42220_full_validation.pdf.gz | 696.6 KB | Display | |
| Data in XML |  emd_42220_validation.xml.gz emd_42220_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_42220_validation.cif.gz emd_42220_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42220 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42220 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42220 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42220 | HTTPS FTP |
-Related structure data
| Related structure data |  8ugbMC  8ufiC  8ugsC  8ulgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42220.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42220.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||
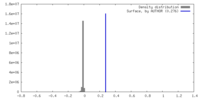
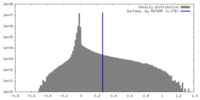
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42220_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
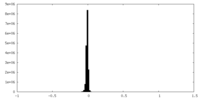
| Density Histograms |
-Half map: #1
| File | emd_42220_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
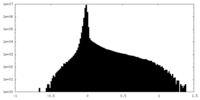
| Density Histograms |
- Sample components
Sample components
-Entire : Bovine rod phosphodiesterase 6 bound to udenafil
| Entire | Name: Bovine rod phosphodiesterase 6 bound to udenafil |
|---|---|
| Components |
|
-Supramolecule #1: Bovine rod phosphodiesterase 6 bound to udenafil
| Supramolecule | Name: Bovine rod phosphodiesterase 6 bound to udenafil / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3, #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha
| Macromolecule | Name: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: 3',5'-cyclic-GMP phosphodiesterase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 99.461789 KDa |
| Sequence | String: MGEVTAEEVE KFLDSNVSFA KQYYNLRYRA KVISDLLGPR EAAVDFSNYH ALNSVEESEI IFDLLRDFQD NLQAEKCVFN VMKKLCFLL QADRMSLFMY RARNGIAELA TRLFNVHKDA VLEECLVAPD SEIVFPLDMG VVGHVALSKK IVNVPNTEED E HFCDFVDT ...String: MGEVTAEEVE KFLDSNVSFA KQYYNLRYRA KVISDLLGPR EAAVDFSNYH ALNSVEESEI IFDLLRDFQD NLQAEKCVFN VMKKLCFLL QADRMSLFMY RARNGIAELA TRLFNVHKDA VLEECLVAPD SEIVFPLDMG VVGHVALSKK IVNVPNTEED E HFCDFVDT LTEYQTKNIL ASPIMNGKDV VAIIMVVNKV DGPHFTENDE EILLKYLNFA NLIMKVFHLS YLHNCETRRG QI LLWSGSK VFEELTDIER QFHKALYTVR AFLNCDRYSV GLLDMTKQKE FFDVWPVLMG EAPPYAGPRT PDGREINFYK VID YILHGK EDIKVIPNPP PDHWALVSGL PTYVAQNGLI CNIMNAPSED FFAFQKEPLD ESGWMIKNVL SMPIVNKKEE IVGV ATFYN RKDGKPFDEM DETLMESLTQ FLGWSVLNPD TYELMNKLEN RKDIFQDMVK YHVKCDNEEI QTILKTREVY GKEPW ECEE EELAEILQGE LPDADKYEIN KFHFSDLPLT ELELVKCGIQ MYYELKVVDK FHIPQEALVR FMYSLSKGYR RITYHN WRH GFNVGQTMFS LLVTGKLKRY FTDLEALAMV TAAFCHDIDH RGTNNLYQMK SQNPLAKLHG SSILERHHLE FGKTLLR DE SLNIFQNLNR RQHEHAIHMM DIAIIATDLA LYFKKRTMFQ KIVDQSKTYE TQQEWTQYMM LDQTRKEIVM AMMMTACD L SAITKPWEVQ SKVALLVAAE FWEQGDLERT VLQQNPIPMM DRNKADELPK LQVGFIDFVC TFVYKEFSRF HEEITPMLD GITNNRKEWK ALADEYETKM KGLEEEKQKQ QAANQAAAGS QHGGKQPGGG PASKSCCVQ UniProtKB: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha |
-Macromolecule #2: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta
| Macromolecule | Name: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: 3',5'-cyclic-GMP phosphodiesterase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 98.449648 KDa |
| Sequence | String: MSLSEGQVHR FLDQNPGFAD QYFGRKLSPE DVANACEDGC PEGCTSFREL CQVEESAALF ELVQDMQENV NMERVVFKIL RRLCSILHA DRCSLFMYRQ RNGVAELATR LFSVQPDSVL EDCLVPPDSE IVFPLDIGVV GHVAQTKKMV NVQDVMECPH F SSFADELT ...String: MSLSEGQVHR FLDQNPGFAD QYFGRKLSPE DVANACEDGC PEGCTSFREL CQVEESAALF ELVQDMQENV NMERVVFKIL RRLCSILHA DRCSLFMYRQ RNGVAELATR LFSVQPDSVL EDCLVPPDSE IVFPLDIGVV GHVAQTKKMV NVQDVMECPH F SSFADELT DYVTRNILAT PIMNGKDVVA VIMAVNKLDG PCFTSEDEDV FLKYLNFGTL NLKIYHLSYL HNCETRRGQV LL WSANKVF EELTDIERQF HKAFYTVRAY LNCDRYSVGL LDMTKEKEFF DVWPVLMGEA QAYSGPRTPD GREILFYKVI DYI LHGKED IKVIPSPPAD HWALASGLPT YVAESGFICN IMNAPADEMF NFQEGPLDDS GWIVKNVLSM PIVNKKEEIV GVAT FYNRK DGKPFDEQDE VLMESLTQFL GWSVLNTDTY DKMNKLENRK DIAQDMVLYH VRCDREEIQL ILPTRERLGK EPADC EEDE LGKILKEVLP GPAKFDIYEF HFSDLECTEL ELVKCGIQMY YELGVVRKFQ IPQEVLVRFL FSVSKGYRRI TYHNWR HGF NVAQTMFTLL MTGKLKSYYT DLEAFAMVTA GLCHDIDHRG TNNLYQMKSQ NPLAKLHGSS ILERHHLEFG KFLLSEE TL NIYQNLNRRQ HEHVIHLMDI AIIATDLALY FKKRTMFQKI VDESKNYEDR KSWVEYLSLE TTRKEIVMAM MMTACDLS A ITKPWEVQSK VALLVAAEFW EQGDLERTVL DQQPIPMMDR NKAAELPKLQ VGFIDFVCTF VYKEFSRFHE EILPMFDRL QNNRKEWKAL ADEYEAKVKA LEEDQKKETT AKKVGTEICN GGPAPRSSTC RIL UniProtKB: Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta |
-Macromolecule #3: Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiestera...
| Macromolecule | Name: Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: 3',5'-cyclic-GMP phosphodiesterase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.684229 KDa |
| Sequence | String: MNLEPPKAEI RSATRVMGGP VTPRKGPPKF KQRQTRQFKS KPPKKGVQGF GDDIPGMEGL GTDITVICPW EAFNHLELHE LAQYGII UniProtKB: Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma |
-Macromolecule #4: CYCLIC GUANOSINE MONOPHOSPHATE
| Macromolecule | Name: CYCLIC GUANOSINE MONOPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: PCG |
|---|---|
| Molecular weight | Theoretical: 345.205 Da |
| Chemical component information |  ChemComp-PCG: |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: Udenafil
| Macromolecule | Name: Udenafil / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZUD |
|---|---|
| Molecular weight | Theoretical: 516.656 Da |
| Chemical component information | 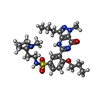 ChemComp-ZUD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 63000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)