[English] 日本語
 Yorodumi
Yorodumi- EMDB-42027: Cryo-EM Structure of Cognate Substrate ATP Bound in the Entry Sit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Cognate Substrate ATP Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | |||||||||
 Map data Map data | Structure of Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondrial RNA Polymerase / Nucleotide Selection / Nucleotide Discrimination / TRANSCRIPTION / Protein-RNA-DNA Complex / POLRMT / TRANSCRIPTION-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtranscription elongation by mitochondrial RNA polymerase / Mitochondrial transcription initiation / positive regulation of mitochondrial transcription / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / regulation of oxidative phosphorylation / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription ...transcription elongation by mitochondrial RNA polymerase / Mitochondrial transcription initiation / positive regulation of mitochondrial transcription / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / regulation of oxidative phosphorylation / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription / transcription elongation factor activity / DNA polymerase processivity factor activity / mitochondrial nucleoid / Transcriptional activation of mitochondrial biogenesis / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / 3'-5'-RNA exonuclease activity / sequence-specific DNA binding / mitochondrial matrix / ribonucleoprotein complex / protein-containing complex / mitochondrion / RNA binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
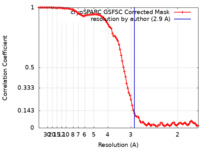
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Herbine KH / Nayak AR / Temiakov D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for substrate binding and selection by human mitochondrial RNA polymerase. Authors: Karl Herbine / Ashok R Nayak / Dmitry Temiakov /  Abstract: The mechanism by which RNAP selects cognate substrates and discriminates between deoxy and ribonucleotides is of fundamental importance to the fidelity of transcription. Here, we present cryo-EM ...The mechanism by which RNAP selects cognate substrates and discriminates between deoxy and ribonucleotides is of fundamental importance to the fidelity of transcription. Here, we present cryo-EM structures of human mitochondrial transcription elongation complexes that reveal substrate ATP bound in Entry and Insertion Sites. In the Entry Site, the substrate binds along the O helix of the fingers domain of mtRNAP but does not interact with the templating DNA base. Interactions between RNAP and the triphosphate moiety of the NTP in the Entry Site ensure discrimination against nucleosides and their diphosphate and monophosphate derivatives but not against non-cognate rNTPs and dNTPs. Closing of the fingers domain over the catalytic site results in delivery of both the templating DNA base and the substrate into the Insertion Site and recruitment of the catalytic magnesium ions. The cryo-EM data also reveal a conformation adopted by mtRNAP to reject a non-cognate substrate from its active site. Our findings establish a structural basis for substrate binding and suggest a unified mechanism of NTP selection for single-subunit RNAPs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42027.map.gz emd_42027.map.gz | 85.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42027-v30.xml emd-42027-v30.xml emd-42027.xml emd-42027.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42027_fsc.xml emd_42027_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42027.png emd_42027.png | 135.5 KB | ||
| Filedesc metadata |  emd-42027.cif.gz emd-42027.cif.gz | 8 KB | ||
| Others |  emd_42027_half_map_1.map.gz emd_42027_half_map_1.map.gz emd_42027_half_map_2.map.gz emd_42027_half_map_2.map.gz | 91.7 MB 91.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42027 http://ftp.pdbj.org/pub/emdb/structures/EMD-42027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42027 | HTTPS FTP |
-Related structure data
| Related structure data |  8u8uMC  8u8vC  9bdcC  9bddC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42027.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42027.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
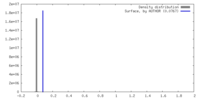
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_42027_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
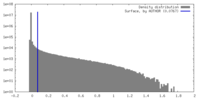
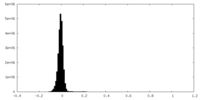
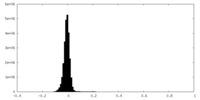
| Density Histograms |
-Half map: Half Map 2
| File | emd_42027_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
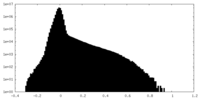
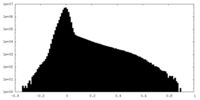
| Density Histograms |
- Sample components
Sample components
-Entire : Cognate Substrate Bound in the Entry Site (ES) of Human Mitochond...
| Entire | Name: Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex |
|---|---|
| Components |
|
-Supramolecule #1: Cognate Substrate Bound in the Entry Site (ES) of Human Mitochond...
| Supramolecule | Name: Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Human Mitochondrial RNA polymerase (d119) and Human TEFM (d50) assembled on an RNA-DNA Scaffold in presence of methylene a,b-ATP. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 227 KDa |
-Macromolecule #1: Transcription elongation factor, mitochondrial
| Macromolecule | Name: Transcription elongation factor, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.262389 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDENAKEPEN RFLRKLLKPD IERERLKAVN SIISIVFGTR RIAWAHLDRK LTVLDWQQSD RWSLMRGIYS SSVYLEEISS IISKMPKAD FYVLEKTGLS IQNSSLFPIL LHFHIMEAML YALLNKTFAQ DGQHQVLSMN RNAVGKHFEL MIGDSRTSGK E LVKQFLFD ...String: MDENAKEPEN RFLRKLLKPD IERERLKAVN SIISIVFGTR RIAWAHLDRK LTVLDWQQSD RWSLMRGIYS SSVYLEEISS IISKMPKAD FYVLEKTGLS IQNSSLFPIL LHFHIMEAML YALLNKTFAQ DGQHQVLSMN RNAVGKHFEL MIGDSRTSGK E LVKQFLFD SILKADPRVF FPSDKIVHYR QMFLSTELQR VEELYDSLLQ AIAFYELAVF DSQPLEHHHH HHHH UniProtKB: Transcription elongation factor, mitochondrial |
-Macromolecule #2: DNA-directed RNA polymerase, mitochondrial
| Macromolecule | Name: DNA-directed RNA polymerase, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.150102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHGR WAKILEKDKR TQQMRMQRLK AKLQMPFQSG EFKALTRRLQ VEPRLLSKQM AGCLEDCTRQ APESPWEEQL ARLLQEAPG KLSLDVEQAP SGQHSQAQLS GQQQRLLAFF KCCLLTDQLP LAHHLLVVHH GQRQKRKLLT LDMYNAVMLG W ARQGAFKE ...String: MGHHHHHHGR WAKILEKDKR TQQMRMQRLK AKLQMPFQSG EFKALTRRLQ VEPRLLSKQM AGCLEDCTRQ APESPWEEQL ARLLQEAPG KLSLDVEQAP SGQHSQAQLS GQQQRLLAFF KCCLLTDQLP LAHHLLVVHH GQRQKRKLLT LDMYNAVMLG W ARQGAFKE LVYVLFMVKD AGLTPDLLSY AAALQCMGRQ DQDAGTIERC LEQMSQEGLK LQALFTAVLL SEEDRATVLK AV HKVKPTF SLPPQLPPPV NTSKLLRDVY AKDGRVSYPK LHLPLKTLQC LFEKQLHMEL ASRVCVVSVE KPTLPSKEVK HAR KTLKTL RDQWEKALCR ALRETKNRLE REVYEGRFSL YPFLCLLDER EVVRMLLQVL QALPAQGESF TTLARELSAR TFSR HVVQR QRVSGQVQAL QNHYRKYLCL LASDAEVPEP CLPRQYWEAL GAPEALREQP WPLPVQMELG KLLAEMLVQA TQMPC SLDK PHRSSRLVPV LYHVYSFRNV QQIGILKPHP AYVQLLEKAA EPTLTFEAVD VPMLCPPLPW TSPHSGAFLL SPTKLM RTV EGATQHQELL ETCPPTALHG ALDALTQLGN CAWRVNGRVL DLVLQLFQAK GCPQLGVPAP PSEAPQPPEA HLPHSAA PA RKAELRRELA HCQKVAREMH SLRAEALYRL SLAQHLRDRV FWLPHNMDFR GRTYPCPPHF NHLGSDVARA LLEFAQGR P LGPHGLDWLK IHLVNLTGLK KREPLRKRLA FAEEVMDDIL DSADQPLTGR KWWMGAEEPW QTLACCMEVA NAVRASDPA AYVSHLPVHQ DGSCNGLQHY AALGRDSVGA ASVNLEPSDV PQDVYSGVAA QVEVFRRQDA QRGMRVAQVL EGFITRKVVK QTVMTVVYG VTRYGGRLQI EKRLRELSDF PQEFVWEASH YLVRQVFKSL QEMFSGTRAI QHWLTESARL ISHMGSVVEW V TPLGVPVI QPYRLDSKVK QIGGGIQSIT YTHNGDISRK PNTRKQKNGF PPNFIHSLDS SHMMLTALHC YRKGLTFVSV HD CYWTHAA DVSVMNQVCR EQFVRLHSEP ILQDLSRFLV KRFCSEPQKI LEASQLKETL QAVPKPGAFD LEQVKRSTYF FS UniProtKB: DNA-directed RNA polymerase, mitochondrial |
-Macromolecule #3: Non-Template Strand DNA (NT27mt)
| Macromolecule | Name: Non-Template Strand DNA (NT27mt) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.483752 KDa |
| Sequence | String: (DG)(DG)(DA)(DC)(DA)(DT)(DG)(DG)(DT)(DG) (DT)(DA)(DA)(DT)(DT)(DA)(DT)(DT)(DT)(DC) (DG)(DA)(DC)(DG)(DC)(DC)(DA)(DG)(DA) (DC)(DG)(DA)(DC)(DC) |
-Macromolecule #5: Template Strand DNA (TS31mt)
| Macromolecule | Name: Template Strand DNA (TS31mt) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.41965 KDa |
| Sequence | String: (DG)(DG)(DT)(DC)(DG)(DT)(DC)(DT)(DG)(DG) (DC)(DG)(DT)(DG)(DC)(DG)(DC)(DG)(DC)(DC) (DG)(DT)(DT)(DA)(DC)(DA)(DC)(DC)(DA) (DT)(DG)(DT)(DC)(DC) |
-Macromolecule #4: RNA (RNA14mt)
| Macromolecule | Name: RNA (RNA14mt) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.493723 KDa |
| Sequence | String: AGUCUGCGGC GCGC |
-Macromolecule #6: DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER
| Macromolecule | Name: DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: APC |
|---|---|
| Molecular weight | Theoretical: 505.208 Da |
| Chemical component information | 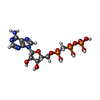 ChemComp-APC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 20 mM Tris Buffer, pH 7.9, 150 mM NaCl, 10 mM DTT, and 5 mM MgCl2 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 15 mA Discharge |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | 5 uM complex of (D119) wild-type mtRNAP, (D50) wild-type TEFM, an RNA-DNA scaffold (R14/T31/NT27), and a,b methylene ATP. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Details | Preliminary grid screening performed manually using TFS Glacios. |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 11439 / Average exposure time: 3.4 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial Fitting was done in Chimera and flexible/refined fitting done with Coot Phenix Real-Space Refinement. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 45 / Target criteria: Cross-correlation Coefficient |
| Output model |  PDB-8u8u: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)