[English] 日本語
 Yorodumi
Yorodumi- EMDB-42004: KCTD5/Cullin3/Gbeta1gamma2 Complex State C: Composite Map from RE... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KCTD5/Cullin3/Gbeta1gamma2 Complex State C: Composite Map from RELION Multi-body Refinement | |||||||||
 Map data Map data | KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Full Mape | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cullin family protein / proteasome-mediated ubiquitin-dependent protein catabolic process / complex / ubiquitin-dependent protein catabolic process / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of mitotic cell cycle phase transition / trophectodermal cellular morphogenesis / liver morphogenesis / POZ domain binding / polar microtubule / nuclear protein quality control by the ubiquitin-proteasome system / regulation protein catabolic process at postsynapse / COPII vesicle coating / anaphase-promoting complex-dependent catabolic process / positive regulation of mitotic metaphase/anaphase transition ...positive regulation of mitotic cell cycle phase transition / trophectodermal cellular morphogenesis / liver morphogenesis / POZ domain binding / polar microtubule / nuclear protein quality control by the ubiquitin-proteasome system / regulation protein catabolic process at postsynapse / COPII vesicle coating / anaphase-promoting complex-dependent catabolic process / positive regulation of mitotic metaphase/anaphase transition / RHOBTB3 ATPase cycle / embryonic cleavage / cell projection organization / Notch binding / fibroblast apoptotic process / RHOBTB1 GTPase cycle / negative regulation of type I interferon production / stem cell division / mitotic metaphase chromosome alignment / Cul3-RING ubiquitin ligase complex / negative regulation of Rho protein signal transduction / stress fiber assembly / positive regulation of cytokinesis / ubiquitin ligase complex scaffold activity / cullin family protein binding / protein monoubiquitination / endoplasmic reticulum to Golgi vesicle-mediated transport / RHOBTB2 GTPase cycle / sperm flagellum / protein K48-linked ubiquitination / protein autoubiquitination / gastrulation / positive regulation of TORC1 signaling / regulation of cellular response to insulin stimulus / intrinsic apoptotic signaling pathway / cyclin binding / positive regulation of protein ubiquitination / integrin-mediated signaling pathway / cellular response to amino acid stimulus / kidney development / Degradation of DVL / G1/S transition of mitotic cell cycle / Hedgehog 'on' state / protein homooligomerization / protein destabilization / Wnt signaling pathway / Olfactory Signaling Pathway / protein polyubiquitination / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / spindle pole / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Regulation of RAS by GAPs / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / mitotic spindle / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / ubiquitin protein ligase activity / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / KEAP1-NFE2L2 pathway / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / cell migration / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / Antigen processing: Ubiquitination & Proteasome degradation / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / Neddylation / gene expression / retina development in camera-type eye / GTPase binding / cellular response to oxidative stress / Ca2+ pathway / fibroblast proliferation / ubiquitin-dependent protein catabolic process Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
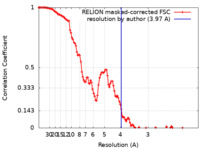
| Method | single particle reconstruction / cryo EM / Resolution: 3.97 Å | |||||||||
 Authors Authors | Nguyen DM / Narayanan N / Kuntz DA / Prive GG | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure and dynamics of a pentameric KCTD5/CUL3/Gβγ E3 ubiquitin ligase complex. Authors: Duc Minh Nguyen / Deanna H Rath / Dominic Devost / Darlaine Pétrin / Robert Rizk / Alan X Ji / Naveen Narayanan / Darren Yong / Andrew Zhai / Douglas A Kuntz / Maha U Q Mian / Neil C Pomroy ...Authors: Duc Minh Nguyen / Deanna H Rath / Dominic Devost / Darlaine Pétrin / Robert Rizk / Alan X Ji / Naveen Narayanan / Darren Yong / Andrew Zhai / Douglas A Kuntz / Maha U Q Mian / Neil C Pomroy / Alexander F A Keszei / Samir Benlekbir / Mohammad T Mazhab-Jafari / John L Rubinstein / Terence E Hébert / Gilbert G Privé /  Abstract: Heterotrimeric G proteins can be regulated by posttranslational modifications, including ubiquitylation. KCTD5, a pentameric substrate receptor protein consisting of an N-terminal BTB domain and a C- ...Heterotrimeric G proteins can be regulated by posttranslational modifications, including ubiquitylation. KCTD5, a pentameric substrate receptor protein consisting of an N-terminal BTB domain and a C-terminal domain, engages CUL3 to form the central scaffold of a cullin-RING E3 ligase complex (CRL3) that ubiquitylates Gβγ and reduces Gβγ protein levels in cells. The cryo-EM structure of a 5:5:5 KCTD5/CUL3/Gβγ assembly reveals a highly dynamic complex with rotations of over 60° between the KCTD5/CUL3 and KCTD5/Gβγ moieties of the structure. CRL3 engages the E3 ligase ARIH1 to ubiquitylate Gβγ in an E3-E3 superassembly, and extension of the structure to include full-length CUL3 with RBX1 and an ARIH1~ubiquitin conjugate reveals that some conformational states position the ARIH1~ubiquitin thioester bond to within 10 Å of lysine-23 of Gβ and likely represent priming complexes. Most previously described CRL/substrate structures have consisted of monovalent complexes and have involved flexible peptide substrates. The structure of the KCTD5/CUL3/Gβγ complex shows that the oligomerization of a substrate receptor can generate a polyvalent E3 ligase complex and that the internal dynamics of the substrate receptor can position a structured target for ubiquitylation in a CRL3 complex. #1:  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Structure and dynamics of a pentameric KCTD5/Cullin3/G beta gamma E3 ubiquitin ligase complex Authors: Nguyen DM / Rath DH / Devost D / Petrin D / Rizk R / Ji AX / Narayanan N / Yong D / Zhai A / Kuntz DA / Mian MUQ / Pomroy NC / Keszei AFE / Benlekbir S / Mazhab-Jafari MT / Rubinstein JL / Hebert TE / Prive GG | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42004.map.gz emd_42004.map.gz | 108 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42004-v30.xml emd-42004-v30.xml emd-42004.xml emd-42004.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42004_fsc.xml emd_42004_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42004.png emd_42004.png | 102.9 KB | ||
| Masks |  emd_42004_msk_1.map emd_42004_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42004.cif.gz emd-42004.cif.gz | 6.2 KB | ||
| Others |  emd_42004_half_map_1.map.gz emd_42004_half_map_1.map.gz emd_42004_half_map_2.map.gz emd_42004_half_map_2.map.gz | 108.3 MB 108.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42004 http://ftp.pdbj.org/pub/emdb/structures/EMD-42004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42004 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42004 | HTTPS FTP |
-Related structure data
| Related structure data |  8u83MC  8u7zC  8u80C  8u81C  8u82C  8u84C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42004.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42004.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Full Mape | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42004_msk_1.map emd_42004_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Half Map...
| File | emd_42004_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Half Map...
| File | emd_42004_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KCTD5:Cullin3:GBetaGamma StateC Composite map from Multibody: Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of substrate adaptor KCTD5 with Cullin-3 and substrate Gb...
| Entire | Name: Complex of substrate adaptor KCTD5 with Cullin-3 and substrate Gbeta-gamma |
|---|---|
| Components |
|
-Supramolecule #1: Complex of substrate adaptor KCTD5 with Cullin-3 and substrate Gb...
| Supramolecule | Name: Complex of substrate adaptor KCTD5 with Cullin-3 and substrate Gbeta-gamma type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: KCTD5_HUMAN
| Macromolecule | Name: KCTD5_HUMAN / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MAENHCELLS PARGGIGAGL GGGLCRRCSA GLGALAQRPG SVSKWVRLNV GGTYFLTTRQ TLCRDPKSF LYRLCQADPD LDSDKDETGA YLIDRDPTYF GPVLNYLRHG KLVINKDLAE E GVLEEAEF YNITSLIKLV KDKIRERDSK TSQVPVKHVY RVLQCQEEEL ...String: MAENHCELLS PARGGIGAGL GGGLCRRCSA GLGALAQRPG SVSKWVRLNV GGTYFLTTRQ TLCRDPKSF LYRLCQADPD LDSDKDETGA YLIDRDPTYF GPVLNYLRHG KLVINKDLAE E GVLEEAEF YNITSLIKLV KDKIRERDSK TSQVPVKHVY RVLQCQEEEL TQMVSTMSDG WK FEQLVSI GSSYNYGNED QAEFLCVVSK ELHNTPYGTA SEPSEKAKIL QERGSRM UniProtKB: BTB/POZ domain-containing protein KCTD5 |
-Macromolecule #2: cullin3 1-381
| Macromolecule | Name: cullin3 1-381 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MSNLSKGTGS RKDTKMRIRA FPMTMDEKYV NSIWDLLKNA IQEIQRKNNS GLSFEELYRN AYTMVLHKHG EKLYTGLREV VTEHLINKVR EDVLNSLNNN FLQTLNQAWN DHQTAMVMIR DILMYMDRVY VQQNNVENVY NLGLIIFRDQ VVRYGCIRDH LRQTLLDMIA ...String: MSNLSKGTGS RKDTKMRIRA FPMTMDEKYV NSIWDLLKNA IQEIQRKNNS GLSFEELYRN AYTMVLHKHG EKLYTGLREV VTEHLINKVR EDVLNSLNNN FLQTLNQAWN DHQTAMVMIR DILMYMDRVY VQQNNVENVY NLGLIIFRDQ VVRYGCIRDH LRQTLLDMIA RERKGEVVDR GAIRNACQML MILGLEGRSV YEEDFEAPFL EMSAEFFQME SQKFLAENSA SVYIKKVEAR INEEIERVMH CLDKSTEEPI VKVVERELIS KHMKTIVEME NSGLVHMLKN GKTEDLGCMY KLFSRVPNGL KTMCECMSSY LREQGKALVS EEGEGKNPVD YIQGLLDLKS RFDRFLLESF NNDRLFKQTI AGDFEYFLNL N UniProtKB: Cullin-3 |
-Macromolecule #3: GBB1_HUMAN
| Macromolecule | Name: GBB1_HUMAN / type: protein_or_peptide / ID: 3 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRL LVSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN I CSIYNLKT REGNVRVSRE LAGHTGYLSC CRFLDDNQIV TSSGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRL LVSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN I CSIYNLKT REGNVRVSRE LAGHTGYLSC CRFLDDNQIV TSSGDTTCAL WDIETGQQTT TF TGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DINAICFFPN GNA FATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDAL KADRA GVLAGHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: GBG2_HUMAN
| Macromolecule | Name: GBG2_HUMAN / type: protein_or_peptide / ID: 4 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSA IL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 7464 / Average electron dose: 49.35 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.25 µm / Nominal magnification: 75000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8u83: |
 Movie
Movie Controller
Controller










































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)