[English] 日本語
 Yorodumi
Yorodumi- EMDB-41947: Locally refined (symmetry based) cryo-EM map of the second dimer ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined (symmetry based) cryo-EM map of the second dimer of the SPARTA oligomer with guide RNA and target DNA | |||||||||
 Map data Map data | locally refined map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA / DNA / Argonaute / TIR / immune system / RNA BINDING PROTEIN | |||||||||
| Biological species |  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Malik R / Kottur J / Aggarwal AK | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Nucleic acid mediated activation of a short prokaryotic Argonaute immune system. Authors: Jithesh Kottur / Radhika Malik / Aneel K Aggarwal /   Abstract: A short prokaryotic Argonaute (pAgo) TIR-APAZ (SPARTA) defense system, activated by invading DNA to unleash its TIR domain for NAD(P) hydrolysis, was recently identified in bacteria. We report the ...A short prokaryotic Argonaute (pAgo) TIR-APAZ (SPARTA) defense system, activated by invading DNA to unleash its TIR domain for NAD(P) hydrolysis, was recently identified in bacteria. We report the crystal structure of SPARTA heterodimer in the absence of guide-RNA/target-ssDNA (2.66 Å) and a cryo-EM structure of the SPARTA oligomer (tetramer of heterodimers) bound to guide-RNA/target-ssDNA at nominal 3.15-3.35 Å resolution. The crystal structure provides a high-resolution view of SPARTA, revealing the APAZ domain as equivalent to the N, L1, and L2 regions of long pAgos and the MID domain containing a unique insertion (insert57). Cryo-EM structure reveals regions of the PIWI (loop10-9) and APAZ (helix αN) domains that reconfigure for nucleic-acid binding and decrypts regions/residues that reorganize to expose a positively charged pocket for higher-order assembly. The TIR domains amass in a parallel-strands arrangement for catalysis. We visualize SPARTA before and after RNA/ssDNA binding and uncover the basis of its active assembly leading to abortive infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41947.map.gz emd_41947.map.gz | 398.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41947-v30.xml emd-41947-v30.xml emd-41947.xml emd-41947.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41947.png emd_41947.png | 85 KB | ||
| Filedesc metadata |  emd-41947.cif.gz emd-41947.cif.gz | 5.1 KB | ||
| Others |  emd_41947_half_map_1.map.gz emd_41947_half_map_1.map.gz emd_41947_half_map_2.map.gz emd_41947_half_map_2.map.gz | 390.8 MB 390.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41947 http://ftp.pdbj.org/pub/emdb/structures/EMD-41947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41947 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41947.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41947.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally refined map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
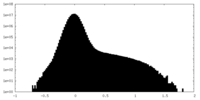
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
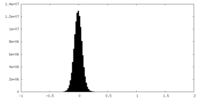
| Density Histograms |
-Half map: half-map
| File | emd_41947_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
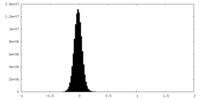
| Density Histograms |
- Sample components
Sample components
-Entire : SPARTA complex with gRNA/tDNA
| Entire | Name: SPARTA complex with gRNA/tDNA |
|---|---|
| Components |
|
-Supramolecule #1: SPARTA complex with gRNA/tDNA
| Supramolecule | Name: SPARTA complex with gRNA/tDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
-Macromolecule #1: Ago
| Macromolecule | Name: Ago / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MKELIYIEEP SILFAHGQKC TDPRDGLALF GPLNQIYGIK SGVVGTQKGL QIFKSYLDKI QKPIYNHNN ITRPMFPGFE AVFGCKWESQ NIVFKEITDE EIRRYLFNAS THKRTYDLVT L FNDKIITA NKNDEERVDV WFVIVPEEIY KYCRPNSVLP NELVQTKSLI ...String: MKELIYIEEP SILFAHGQKC TDPRDGLALF GPLNQIYGIK SGVVGTQKGL QIFKSYLDKI QKPIYNHNN ITRPMFPGFE AVFGCKWESQ NIVFKEITDE EIRRYLFNAS THKRTYDLVT L FNDKIITA NKNDEERVDV WFVIVPEEIY KYCRPNSVLP NELVQTKSLI SKSKAKSFRY TP TLFEEFN KKLKEVEKEA KTYNYDAQFH DQLKARLLEH TIPTQILRES TLAWRDFKNT FGA PIRDFS KIEGHLAWTI STAAYYKAGG KPWKLGDIRP GVCYLGLVYK KIEKSKNPQN ACCA AQMFL DNGDGTVFKG EVGPWYNPEK GEYHLKPKEA KALLTQALES YKEQNKSYPK EVFIH ARTR FNDEEWNAFN EVTPKNTNLV GVTITKSKPL KLYKTEGAFP IMRGNAYIVD EKKAFL WTL GFVPKLQSTL SMEVPNPIFI EINKGEAEIQ QVLKDILALT KLNYNACIYA DGEPVTL RF ANKIGEILTA STEIKTPPLA FKYYI |
-Macromolecule #2: TIR-APAZ
| Macromolecule | Name: TIR-APAZ / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermoflavifilum thermophilum (bacteria) Thermoflavifilum thermophilum (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MRNKIFISHA TPEDDDFTRW LSLKLIGLGY EVWCDILFLD KGVDFWSTIE KEIRENTCKF LIVSSTAGN KREGVLKELA VATKVKKHLQ DDMFIIPLAI DENLSYDDIN IEIVRLNAID F KKSWAKGL QDLLDAFEKQ NVPKKPPDHS KSNLLYQQIF LHDKQAIEKE ...String: MRNKIFISHA TPEDDDFTRW LSLKLIGLGY EVWCDILFLD KGVDFWSTIE KEIRENTCKF LIVSSTAGN KREGVLKELA VATKVKKHLQ DDMFIIPLAI DENLSYDDIN IEIVRLNAID F KKSWAKGL QDLLDAFEKQ NVPKKPPDHS KSNLLYQQIF LHDKQAIEKE ETYDSNWFPI IS FPNELRF HRYDWRLPKQ FDVRTLAFPA IRYKEYLCTF AWEYDFIHQL PKTETYNGQE SIR ISTSDI LSGRYDTDFI RNYECQRLIV QLINKAFELR MKDKNVREYQ MSKTFAYWIE KGKL EKDKF EKIKLVGKQK NKYWHFGISA AGKLYPSPVL MVSSHIIFTM DGINLIKSKS IQHSS RRKQ GKNWWNDKWR EKLLAFIRFL SDDQNAIYLN VGSEEKILIS NKPLKFFGKM SYVTPS EVT LEEESVLADI NNFEEDTEDL DELEDIE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Sugar embedding | Material: Vitreous ice |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 51.11 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.17 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 238432 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)