[English] 日本語
 Yorodumi
Yorodumi- EMDB-41874: CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06 | |||||||||
 Map data Map data | CryoEM map of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza virus / hemagglutinin / H1 / monoclonal antibody / virus / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
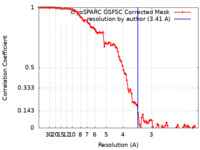
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Moore N / Han J / Ward AB / Wilson IA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Exp Med / Year: 2024 Journal: J Exp Med / Year: 2024Title: Maturation of germinal center B cells after influenza virus vaccination in humans. Authors: Katherine M McIntire / Hailong Meng / Ting-Hui Lin / Wooseob Kim / Nina E Moore / Julianna Han / Meagan McMahon / Meng Wang / Sameer Kumar Malladi / Bassem M Mohammed / Julian Q Zhou / Aaron ...Authors: Katherine M McIntire / Hailong Meng / Ting-Hui Lin / Wooseob Kim / Nina E Moore / Julianna Han / Meagan McMahon / Meng Wang / Sameer Kumar Malladi / Bassem M Mohammed / Julian Q Zhou / Aaron J Schmitz / Kenneth B Hoehn / Juan Manuel Carreño / Temima Yellin / Teresa Suessen / William D Middleton / Sharlene A Teefey / Rachel M Presti / Florian Krammer / Jackson S Turner / Andrew B Ward / Ian A Wilson / Steven H Kleinstein / Ali H Ellebedy /   Abstract: Germinal centers (GC) are microanatomical lymphoid structures where affinity-matured memory B cells and long-lived bone marrow plasma cells are primarily generated. It is unclear how the maturation ...Germinal centers (GC) are microanatomical lymphoid structures where affinity-matured memory B cells and long-lived bone marrow plasma cells are primarily generated. It is unclear how the maturation of B cells within the GC impacts the breadth and durability of B cell responses to influenza vaccination in humans. We used fine needle aspiration of draining lymph nodes to longitudinally track antigen-specific GC B cell responses to seasonal influenza vaccination. Antigen-specific GC B cells persisted for at least 13 wk after vaccination in two out of seven individuals. Monoclonal antibodies (mAbs) derived from persisting GC B cell clones exhibit enhanced binding affinity and breadth to influenza hemagglutinin (HA) antigens compared with related GC clonotypes isolated earlier in the response. Structural studies of early and late GC-derived mAbs from one clonal lineage in complex with H1 and H5 HAs revealed an altered binding footprint. Our study shows that inducing sustained GC reactions after influenza vaccination in humans supports the maturation of responding B cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41874.map.gz emd_41874.map.gz | 86.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41874-v30.xml emd-41874-v30.xml emd-41874.xml emd-41874.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41874_fsc.xml emd_41874_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41874.png emd_41874.png | 88.1 KB | ||
| Masks |  emd_41874_msk_1.map emd_41874_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41874.cif.gz emd-41874.cif.gz | 7.8 KB | ||
| Others |  emd_41874_half_map_1.map.gz emd_41874_half_map_1.map.gz emd_41874_half_map_2.map.gz emd_41874_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41874 http://ftp.pdbj.org/pub/emdb/structures/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41874 | HTTPS FTP |
-Validation report
| Summary document |  emd_41874_validation.pdf.gz emd_41874_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41874_full_validation.pdf.gz emd_41874_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_41874_validation.xml.gz emd_41874_validation.xml.gz | 17.4 KB | Display | |
| Data in CIF |  emd_41874_validation.cif.gz emd_41874_validation.cif.gz | 22.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 | HTTPS FTP |
-Related structure data
| Related structure data |  8u44MC  8txmC  8txpC  8txtC  8ty7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41874.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41874.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41874_msk_1.map emd_41874_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
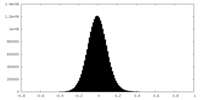
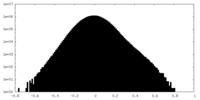
| Density Histograms |
-Half map: CryoEM half map A of A/Solomon Islands/3/2006 H1...
| File | emd_41874_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map A of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM half map B of A/Solomon Islands/3/2006 H1...
| File | emd_41874_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map B of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10
| Entire | Name: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10 |
|---|---|
| Components |
|
-Supramolecule #1: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10
| Supramolecule | Name: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4, #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Fab
| Supramolecule | Name: Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: HA
| Supramolecule | Name: HA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 05.GC.w2.3C10-H1_SI06 Heavy chain
| Macromolecule | Name: 05.GC.w2.3C10-H1_SI06 Heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.771057 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DEVQLVQSGA EVKKPGSSVR VSCKASGGTF SAISWVRQAP GQGLEWMGGI IPVFGTANYA QKFQGRVTI TADDSTSTAY MEVSSLRSDD TAVYYCAREE TWKGATIGVM GIWGQGTMVT VSSASTKGPS VFPLAPSSKS T SGGTAALG ...String: METDTLLLWV LLLWVPGSTG DEVQLVQSGA EVKKPGSSVR VSCKASGGTF SAISWVRQAP GQGLEWMGGI IPVFGTANYA QKFQGRVTI TADDSTSTAY MEVSSLRSDD TAVYYCAREE TWKGATIGVM GIWGQGTMVT VSSASTKGPS VFPLAPSSKS T SGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RV EPKSC |
-Macromolecule #2: 05.GC.w2.3C10-H1_SI06 Light chain
| Macromolecule | Name: 05.GC.w2.3C10-H1_SI06 Light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.706625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DDIQMTQSPS SLSASVGDRV TITCRASQSI SSYLNWYQHK PGKAPKLLIF TASNLQSGVP SRFSGGGSG TDFTLTISSL QPEDFATYYC QQSYTSPRTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN F YPREAKVQ ...String: METDTLLLWV LLLWVPGSTG DDIQMTQSPS SLSASVGDRV TITCRASQSI SSYLNWYQHK PGKAPKLLIF TASNLQSGVP SRFSGGGSG TDFTLTISSL QPEDFATYYC QQSYTSPRTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN F YPREAKVQ WKVDNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #3: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 40.835902 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAAD PGDTICIGYH ANNSTDTVDT VLEKNVTVTH SVNLLEDSHN GKLCRLKGI APLQLGNCSV AGWILGNPEC ELLISRESWS YIVEKPNPEN GTCYPGHFAD YEELREQLSS VSSFERFEIF P KESSWPNH ...String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAAD PGDTICIGYH ANNSTDTVDT VLEKNVTVTH SVNLLEDSHN GKLCRLKGI APLQLGNCSV AGWILGNPEC ELLISRESWS YIVEKPNPEN GTCYPGHFAD YEELREQLSS VSSFERFEIF P KESSWPNH TTTGVSASCS HNGESSFYKN LLWLTGKNGL YPNLSKSYAN NKEKEVLVLW GVHHPPNIGD QRALYHKENA YV SVVSSHY SRKFTPEIAK RPKVRDQEGR INYYWTLLEP GDTIIFEANG NLIAPRYAFA LSRGFGSGII NSNAPMDECD AKC QTPQGA INSSLPFQNV HPVTIGECPK YVRSAKLRMV TGLRNIPSIQ SR UniProtKB: Hemagglutinin |
-Macromolecule #4: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 26.988895 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFI DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNDECM ESVKNGTYDY P KYSEESKL ...String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFI DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNDECM ESVKNGTYDY P KYSEESKL NREKIDSGGG GLNDIFEAQK IEWHERLVPR GSPGSGYIPE APRDGQAYVR KDGEWVLLST FLGHHHHHH UniProtKB: Hemagglutinin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: Tris-buffered saline |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 25 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


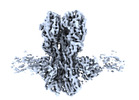






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)