+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Apo Sialin at pH7.5 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Transporter / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsialic acid:proton symporter activity / D-glucuronate transmembrane transporter activity / Defective SLC17A5 causes Salla disease (SD) and ISSD / Organic anion transport by SLC5/17/25 transporters / sialic acid transmembrane transporter activity / sialic acid transport / carbohydrate:proton symporter activity / Sialic acid metabolism / neurotransmitter loading into synaptic vesicle / monoatomic anion transport ...sialic acid:proton symporter activity / D-glucuronate transmembrane transporter activity / Defective SLC17A5 causes Salla disease (SD) and ISSD / Organic anion transport by SLC5/17/25 transporters / sialic acid transmembrane transporter activity / sialic acid transport / carbohydrate:proton symporter activity / Sialic acid metabolism / neurotransmitter loading into synaptic vesicle / monoatomic anion transport / amino acid transport / monoatomic ion transport / response to bacterium / synaptic vesicle membrane / basolateral plasma membrane / lysosome / apical plasma membrane / lysosomal membrane / glutamatergic synapse / membrane / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | ||||||||||||
 Authors Authors | Schmiege P / Li X | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure and inhibition of the human lysosomal transporter Sialin. Authors: Philip Schmiege / Linda Donnelly / Nadia Elghobashi-Meinhardt / Chia-Hsueh Lee / Xiaochun Li /   Abstract: Sialin, a member of the solute carrier 17 (SLC17) transporter family, is unique in its ability to transport not only sialic acid using a pH-driven mechanism, but also transport mono and diacidic ...Sialin, a member of the solute carrier 17 (SLC17) transporter family, is unique in its ability to transport not only sialic acid using a pH-driven mechanism, but also transport mono and diacidic neurotransmitters, such as glutamate and N-acetylaspartylglutamate (NAAG), into synaptic vesicles via a membrane potential-driven mechanism. While most transporters utilize one of these mechanisms, the structural basis of how Sialin transports substrates using both remains unclear. Here, we present the cryogenic electron-microscopy structures of human Sialin: apo cytosol-open, apo lumen-open, NAAG-bound, and inhibitor-bound. Our structures show that a positively charged cytosol-open vestibule accommodates either NAAG or the Sialin inhibitor Fmoc-Leu-OH, while its luminal cavity potentially binds sialic acid. Moreover, functional analyses along with molecular dynamics simulations identify key residues in binding sialic acid and NAAG. Thus, our findings uncover the essential conformational states in NAAG and sialic acid transport, demonstrating a working model of SLC17 transporters. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41858.map.gz emd_41858.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41858-v30.xml emd-41858-v30.xml emd-41858.xml emd-41858.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41858.png emd_41858.png | 27.9 KB | ||
| Filedesc metadata |  emd-41858.cif.gz emd-41858.cif.gz | 5.3 KB | ||
| Others |  emd_41858_half_map_1.map.gz emd_41858_half_map_1.map.gz emd_41858_half_map_2.map.gz emd_41858_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41858 http://ftp.pdbj.org/pub/emdb/structures/EMD-41858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41858 | HTTPS FTP |
-Related structure data
| Related structure data |  8u3dMC  8u3eC  8u3fC  8u3gC  8u3hC  9aybC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41858.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41858.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
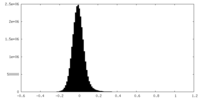
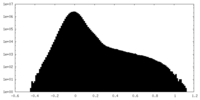
| Density Histograms |
-Half map: #1
| File | emd_41858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
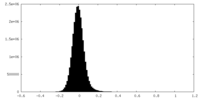
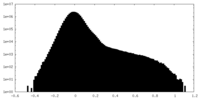
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of Apo Sialin at pH7.5
| Entire | Name: Structure of Apo Sialin at pH7.5 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of Apo Sialin at pH7.5
| Supramolecule | Name: Structure of Apo Sialin at pH7.5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sialin
| Macromolecule | Name: Sialin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.681137 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKR SPVRDLARND GEESTDRTPL LPGAPRAEAA PVCCSARYNL AILAFFGFFI VYALRVNLSV ALVDMVDSNT TLEDNRTSK ACPEHSAPIK VHHNQTGKKY QWDAETQGWI LGSFFYGYII TQIPGGYVAS KIGGKMLLGF GILGTAVLTL F TPIAADLG ...String: MDYKDDDDKR SPVRDLARND GEESTDRTPL LPGAPRAEAA PVCCSARYNL AILAFFGFFI VYALRVNLSV ALVDMVDSNT TLEDNRTSK ACPEHSAPIK VHHNQTGKKY QWDAETQGWI LGSFFYGYII TQIPGGYVAS KIGGKMLLGF GILGTAVLTL F TPIAADLG VGPLIVLRAL EGLGEGVTFP AMHAMWSSWA PPLERSKLLS ISYAGAQLGT VISLPLSGII CYYMNWTYVF YF FGTIGIF WFLLWIWLVS DTPQKHKRIS HYEKEYILSS LRNQLSSQKS VPWVPILKSL PLWAIVVAHF SYNWTFYTLL TLL PTYMKE ILRFNVQENG FLSSLPYLGS WLCMILSGQA ADNLRAKWNF STLCVRRIFS LIGMIGPAVF LVAAGFIGCD YSLA VAFLT ISTTLGGFCS SGFSINHLDI APSYAGILLG ITNTFATIPG MVGPVIAKSL TPDNTVGEWQ TVFYIAAAIN VFGAI FFTL FAKGEVQNWA LNDHHGHRH UniProtKB: Sialin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.83 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 200101 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)