+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C-terminal LRRK2 bound to E11 DARPin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / complex / DARPins / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly ...caveola neck / : / beta-catenin destruction complex binding / regulation of branching morphogenesis of a nerve / Wnt signalosome assembly / regulation of kidney size / regulation of cell projection organization / tangential migration from the subventricular zone to the olfactory bulb / GTP-dependent protein kinase activity / regulation of SNARE complex assembly / regulation of neuroblast proliferation / regulation of ER to Golgi vesicle-mediated transport / protein localization to endoplasmic reticulum exit site / peroxidase inhibitor activity / negative regulation of late endosome to lysosome transport / regulation of mitochondrial depolarization / : / positive regulation of dopamine receptor signaling pathway / amphisome / regulation of synaptic vesicle transport / regulation of lysosomal lumen pH / regulation of CAMKK-AMPK signaling cascade / co-receptor binding / negative regulation of GTPase activity / regulation of dopamine receptor signaling pathway / positive regulation of microglial cell activation / regulation of neuron maturation / regulation of retrograde transport, endosome to Golgi / positive regulation of synaptic vesicle endocytosis / cytoplasmic side of mitochondrial outer membrane / negative regulation of autophagosome assembly / olfactory bulb development / JUN kinase kinase kinase activity / regulation of cAMP/PKA signal transduction / neuron projection arborization / striatum development / multivesicular body, internal vesicle / negative regulation of excitatory postsynaptic potential / regulation of dendritic spine morphogenesis / mitochondrion localization / protein localization to mitochondrion / cellular response to dopamine / positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway / endoplasmic reticulum organization / positive regulation of protein autoubiquitination / negative regulation of protein processing / Wnt signalosome / positive regulation of programmed cell death / GTP metabolic process / regulation of canonical Wnt signaling pathway / syntaxin-1 binding / regulation of reactive oxygen species metabolic process / lysosome organization / Golgi-associated vesicle / clathrin binding / PTK6 promotes HIF1A stabilization / negative regulation of macroautophagy / regulation of mitochondrial fission / regulation of locomotion / protein kinase A binding / neuromuscular junction development / regulation of synaptic vesicle exocytosis / intracellular distribution of mitochondria / Golgi organization / microvillus / exploration behavior / endoplasmic reticulum exit site / autolysosome / locomotory exploration behavior / negative regulation of Notch signaling pathway / MAP kinase kinase kinase activity / canonical Wnt signaling pathway / regulation of synaptic vesicle endocytosis / regulation of synaptic transmission, glutamatergic / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / Rho protein signal transduction / presynaptic cytosol / neuron projection morphogenesis / phagocytic vesicle / cellular response to manganese ion / JNK cascade / positive regulation of autophagy / dendrite cytoplasm / tubulin binding / GTPase activator activity / cellular response to starvation / positive regulation of protein ubiquitination / SNARE binding / determination of adult lifespan / cellular response to reactive oxygen species / mitochondrion organization / excitatory postsynaptic potential / regulation of membrane potential / trans-Golgi network / calcium-mediated signaling / regulation of protein stability / regulation of autophagy / autophagy / small GTPase binding / mitochondrial membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
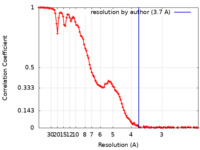
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Sanz-Murillo M / Mathea S / Dederer V / Knapp S / Leschziner A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2024 Journal: J Biol Chem / Year: 2024Title: A designed ankyrin-repeat protein that targets Parkinson's disease-associated LRRK2. Authors: Verena Dederer / Marta Sanz Murillo / Eva P Karasmanis / Kathryn S Hatch / Deep Chatterjee / Franziska Preuss / Kamal R Abdul Azeez / Landon Vu Nguyen / Christian Galicia / Birgit Dreier / ...Authors: Verena Dederer / Marta Sanz Murillo / Eva P Karasmanis / Kathryn S Hatch / Deep Chatterjee / Franziska Preuss / Kamal R Abdul Azeez / Landon Vu Nguyen / Christian Galicia / Birgit Dreier / Andreas Plückthun / Wim Versees / Sebastian Mathea / Andres E Leschziner / Samara L Reck-Peterson / Stefan Knapp /     Abstract: Leucine rich repeat kinase 2 (LRRK2) is a large multidomain protein containing two catalytic domains, a kinase and a GTPase, as well as protein interactions domains, including a WD40 domain. The ...Leucine rich repeat kinase 2 (LRRK2) is a large multidomain protein containing two catalytic domains, a kinase and a GTPase, as well as protein interactions domains, including a WD40 domain. The association of increased LRRK2 kinase activity with both the familial and sporadic forms of Parkinson's disease has led to an intense interest in determining its cellular function. However, small molecule probes that can bind to LRRK2 and report on or affect its cellular activity are needed. Here, we report the identification and characterization of the first high-affinity LRRK2-binding designed ankyrin-repeat protein (DARPin), named E11. Using cryo-EM, we show that DARPin E11 binds to the LRRK2 WD40 domain. LRRK2 bound to DARPin E11 showed improved behavior on cryo-EM grids, resulting in higher resolution LRRK2 structures. DARPin E11 did not affect the catalytic activity of a truncated form of LRRK2 in vitro but decreased the phosphorylation of Rab8A, a LRRK2 substrate, in cells. We also found that DARPin E11 disrupts the formation of microtubule-associated LRRK2 filaments in cells, which are known to require WD40-based dimerization. Thus, DARPin E11 is a new tool to explore the function and dysfunction of LRRK2 and guide the development of LRRK2 kinase inhibitors that target the WD40 domain instead of the kinase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41806.map.gz emd_41806.map.gz | 84.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41806-v30.xml emd-41806-v30.xml emd-41806.xml emd-41806.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41806_fsc.xml emd_41806_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_41806.png emd_41806.png | 40.5 KB | ||
| Filedesc metadata |  emd-41806.cif.gz emd-41806.cif.gz | 7.4 KB | ||
| Others |  emd_41806_half_map_1.map.gz emd_41806_half_map_1.map.gz emd_41806_half_map_2.map.gz emd_41806_half_map_2.map.gz | 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41806 http://ftp.pdbj.org/pub/emdb/structures/EMD-41806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41806 | HTTPS FTP |
-Related structure data
| Related structure data |  8u1bMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41806.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41806.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : C-terminal LRRK2 bound to E11 DARPin
| Entire | Name: C-terminal LRRK2 bound to E11 DARPin |
|---|---|
| Components |
|
-Supramolecule #1: C-terminal LRRK2 bound to E11 DARPin
| Supramolecule | Name: C-terminal LRRK2 bound to E11 DARPin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 156 KDa |
-Macromolecule #1: Leucine-rich repeat serine/threonine-protein kinase 2
| Macromolecule | Name: Leucine-rich repeat serine/threonine-protein kinase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 136.060656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RMKLMIVGNT GSGKTTLLQQ LMKTKKSDLG MQSATVGIDV KDWPIQIRDK RKRDLVLNVW DFAGREEFYS THPHFMTQRA LYLAVYDLS KGQAEVDAMK PWLFNIKARA SSSPVILVGT HLDVSDEKQR KACMSKITKE LLNKRGFPAI RDYHFVNATE E SDALAKLR ...String: RMKLMIVGNT GSGKTTLLQQ LMKTKKSDLG MQSATVGIDV KDWPIQIRDK RKRDLVLNVW DFAGREEFYS THPHFMTQRA LYLAVYDLS KGQAEVDAMK PWLFNIKARA SSSPVILVGT HLDVSDEKQR KACMSKITKE LLNKRGFPAI RDYHFVNATE E SDALAKLR KTIINESLNF KIRDQLVVGQ LIPDCYVELE KIILSERKNV PIEFPVIDRK RLLQLVRENQ LQLDENELPH AV HFLNESG VLLHFQDPAL QLSDLYFVEP KWLCKIMAQI LTVKVEGCPK HPKGIISRRD VEKFLSKKRK FPKNYMSQYF KLL EKFQIA LPIGEEYLLV PSSLSDHRPV IELPHCENSE IIIRLYEMPY FPMGFWSRLI NRLLEISPYM LSGRERALRP NRMY WRQGI YLNWSPEAYC LVGSEVLDNH PESFLKITVP SCRKGCILLG QVVDHIDSLM EEWFPGLLEI DICGEGETLL KKWAL YSFN DGEEHQKILL DDLMKKAEEG DLLVNPDQPR LTIPISQIAP DLILADLPRN IMLNNDELEF EQAPEFLLGD GSFGSV YRA AYEGEEVAVK IFNKHTSLRL LRQELVVLCH LHHPSLISLL AAGIRPRMLV MELASKGSLD RLLQQDKASL TRTLQHR IA LHVADGLRYL HSAMIIYRDL KPHNVLLFTL YPNAAIIAKI ADYGIAQYCC RMGIKTSEGT PGFRAPEVAR GNVIYNQQ A DVYSFGLLLY DILTTGGRIV EGLKFPNEFD ELEIQGKLPD PVKEYGCAPW PMVEKLIKQC LKENPQERPT SAQVFDILN SAELVCLTRR ILLPKNVIVE CMVATHHNSR NASIWLGCGH TDRGQLSFLD LNTEGYTSEE VADSRILCLA LVHLPVEKES WIVSGTQSG TLLVINTEDG KKRHTLEKMT DSVTCLYCNS FSKQSKQKNF LLVGTADGKL AIFEDKTVKL KGAAPLKILN I GNVSTPLM CLSESTNSTE RNVMWGGCGT KIFSFSNDFT IQKLIETRTS QLFSYAAFSD SNIITVVVDT ALYIAKQNSP VV EVWDKKT EKLCGLIDCV HFLREVMVKE NKESKHKMSY SGRVKTLCLQ KNTALWIGTG GGHILLLDLS TRRLIRVIYN FCN SVRVMM TAQLGSLKNV MLVLGYNRKN TEGTQKQKEI QSCLTVWDIN LPHEVQNLEK HIEVRKELAE KMRRTSVE UniProtKB: Leucine-rich repeat serine/threonine-protein kinase 2 |
-Macromolecule #2: E11 DARPin
| Macromolecule | Name: E11 DARPin / type: protein_or_peptide / ID: 2 Details: Nterminal corresponds to HisTag. Some amino acids are not modeled due to the lack of density Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 19.766912 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRGSHHHHHH HHGSDLGKKL LEAARAGQDD EVRILMANGA DVNATDEAGV TPLHLAADSG HLEIVEVLLK TGADVNAWDH YGFTPLHLA AHVGHLEIVE VLLKAGADVN AQDHAGWTPL HLAALYGHLE IVEVLLKHGA DVNAQDMWGE TPFDLAIDNG N EDIAEVLQ KAAKLNDYKD DDDK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.68 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2468 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)