[English] 日本語
 Yorodumi
Yorodumi- EMDB-41680: Characterization of the Chlamydomonas Flagellar Mastigoneme Filam... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of the Chlamydomonas Flagellar Mastigoneme Filament Subunit MST1 Structure at 3.9 angstrom | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fibrous Flagellar Hairs / STRUCTURAL PROTEIN | |||||||||
| Function / homology | Tyrosine-protein kinase ephrin type A/B receptor-like / Tyrosine-protein kinase ephrin type A/B receptor-like / Putative ephrin-receptor like / Growth factor receptor cysteine-rich domain superfamily / Mastigoneme-like protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
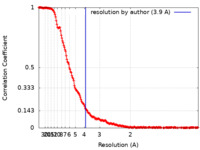
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Yue W / Kai Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2023 Journal: J Cell Biol / Year: 2023Title: Cryo-EM reveals how the mastigoneme assembles and responds to environmental signal changes. Authors: Yue Wang / Jun Yang / Fangheng Hu / Yuchen Yang / Kaiyao Huang / Kai Zhang /   Abstract: Mastigonemes are thread-like structures adorning the flagella of protists. In Chlamydomonas reinhardtii, filamentous mastigonemes find their roots in the flagella's distal region, associated with the ...Mastigonemes are thread-like structures adorning the flagella of protists. In Chlamydomonas reinhardtii, filamentous mastigonemes find their roots in the flagella's distal region, associated with the channel protein PKD2, implying their potential contribution to external signal sensing and flagellar motility control. Here, we present the single-particle cryo-electron microscopy structure of the mastigoneme at 3.4 Å. The filament unit, MST1, consists of nine immunoglobulin-like domains and six Sushi domains, trailed by an elastic polyproline-II helix. Our structure demonstrates that MST1 subunits are periodically assembled to form a centrosymmetric, non-polar filament. Intriguingly, numerous clustered disulfide bonds within a ladder-like spiral configuration underscore structural resilience. While defects in the mastigoneme structure did not noticeably affect general attributes of cell swimming, they did impact specific swimming properties, particularly under varied environmental conditions such as redox shifts and heightened viscosity. Our findings illuminate the potential role of mastigonemes in flagellar motility and suggest their involvement in diverse environmental responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41680.map.gz emd_41680.map.gz | 399.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41680-v30.xml emd-41680-v30.xml emd-41680.xml emd-41680.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41680_fsc.xml emd_41680_fsc.xml | 23.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_41680.png emd_41680.png | 40.7 KB | ||
| Filedesc metadata |  emd-41680.cif.gz emd-41680.cif.gz | 6.4 KB | ||
| Others |  emd_41680_half_map_1.map.gz emd_41680_half_map_1.map.gz emd_41680_half_map_2.map.gz emd_41680_half_map_2.map.gz | 474.9 MB 474.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41680 http://ftp.pdbj.org/pub/emdb/structures/EMD-41680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41680 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41680 | HTTPS FTP |
-Validation report
| Summary document |  emd_41680_validation.pdf.gz emd_41680_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41680_full_validation.pdf.gz emd_41680_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_41680_validation.xml.gz emd_41680_validation.xml.gz | 26.5 KB | Display | |
| Data in CIF |  emd_41680_validation.cif.gz emd_41680_validation.cif.gz | 34.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41680 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41680 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41680 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41680 | HTTPS FTP |
-Related structure data
| Related structure data |  8txcMC  8tx1C  8txbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41680.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41680.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.149 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41680_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
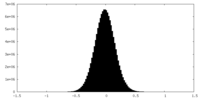
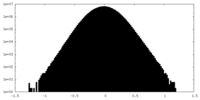
| Density Histograms |
-Half map: #1
| File | emd_41680_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
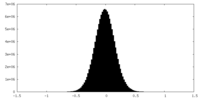
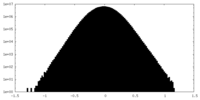
| Density Histograms |
- Sample components
Sample components
-Entire : Mastigoneme
| Entire | Name: Mastigoneme |
|---|---|
| Components |
|
-Supramolecule #1: Mastigoneme
| Supramolecule | Name: Mastigoneme / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Fibrous Flagellar Hairs of Chlamydomonas reinhardtii |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mastigoneme-like protein
| Macromolecule | Name: Mastigoneme-like protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 205.404203 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAMPRHGRRP ARCSSNRASQ WLLLVLGVAL AASPRFLLLV EAATYTLTLD KLGPTVNPTT SDAVTFTATV ASPDSTTAVF FTLDYGDGV TAETTRTTTG ALSTTPTANL VSAGTYTVTY ASIGTKFVTL RLYDSAVAPG VLLASKTVPI YVEDSTLTAT L LQSGVPRL ...String: MAMPRHGRRP ARCSSNRASQ WLLLVLGVAL AASPRFLLLV EAATYTLTLD KLGPTVNPTT SDAVTFTATV ASPDSTTAVF FTLDYGDGV TAETTRTTTG ALSTTPTANL VSAGTYTVTY ASIGTKFVTL RLYDSAVAPG VLLASKTVPI YVEDSTLTAT L LQSGVPRL NLAFSGFKGR VSSSTANRAD MWATIQLDTA PGVFESSRIF IGIAPTASTN YDFVIPDQVY NLEGAKTTVL RI YDAPVGG TLLRTFTPAA ANAVYVVDPS KYVLTLTVGP TSVTTADQVT FTQTTTEVSY SASATSPILQ WRFNWDDPSV VET PLAYPD ALTAASNFPT TATAVSSAAA STFRYTSTGS KNARLRLYDG ANNVIAEKIV VITVSNAGYT LALAKTTADP VTTD DTIAF SAGAKHLSST SQVWWTIDYG AGESSPRTAL TMTNVGAAAP NAIASLSNQY TSGGTKLATL RIYDRDGVGA NTGLL LAST TVTFTVTPVL YALESAVEPF SPIATVAAKW SFRIQRSKAT PAGVTESIKC AFFGADTGTA PADLAAWLTA ANGAGG LTA TILPSSIPSD IISFTRTYAA AAASLQGKLQ CFIGSTPLWD PYYPTPVFQV LAAAPTYTLS ASVTPAVVPV DTATLWT YN IIRSVPVPAG GPSLPILCSF WDGKTGAAPT TDAGWAALAG SANGKGTSMA PGSTTATCSF TPSYSTTGTA TPTLQLIQ N SFALDAATTV GFLSPVYTAP AFATVTAASY TISSYLNPVT PVAGGAAAVW RIVITRNAAV TASAKTLTCQ MPDNGQGGS PADVTADIAV GGTTTVCVFS IAGYTTATPG PYFATVNVVD GAVTTSHITK NFTVLASGTT APTYAVTSVV SPATPVKVST PVTYTFTIT RTTAVPAGGI PQPIICEFFN GEGTAPASAA AYWRVSTTIP DADTVVAVMA PGETTTTCTF TTYYTTVSAG G FTAKLMVF GESATAAPLL TSLSVTPSQL LAAVHSFATP MVVAAAVVAV ESTTISPNYN PTTPYTNIPT YFTFTLLRDP PV PPSASSG VQFACALYTG QNVNPASAPS AITDAVYKTF TDVTTAVATD ANYFADQQLR VVTMAPGTGR VSCTFPTLYA AAG PFSPKF FVFEYASSTV GANALAVADT VTSLTSFTTQ AAPTFITGPT NVPQRVPLPK GFRTTCFDGY ELIFSNDNYT NGVR VAVDA YPYPVGQCRK CPGGTATMDG YRCIPCPSGY WSNEGARECT ACPAGTIAKP AALTARAKYS IDPTTYHFVT HLAMG PESC KKCPKGYFQP NIAGTVCLPC PSGFVSTSGA TGCTACSEGT YHTDGVGTTT PGEATSLDTT DTFGSIYPII PNTCRQ CPA NTYLPLRGQA AIASMNLAAV SSATPCRPCE DGTWSKAGAA GCQKCPPGTY RNTWFSGQLG SPFITADGVP VATTLTE LG SGCSQCPPGT YAPTFGMSVC LPCPAGTFAS APGATACQQC KPGTNSLMGD RTQQMALVVT NAANDFPALR AYTISGMV A GPAYAKPIVT GPDTNFFMAG KSETCSTNLP GYYTDVDGLP IQLPCKPGTF MPFDTATANL LDTGLTVDGT QCYTCQTGT FNDEFSQPVC KACWSGSFAS KRGLPTCEIA QPGTFTNVAA AANATFNTAT LIPTGLVKGA QAPTPCGMGY FQSSAETTTC TACAVGTYA DQAGLAACKP CQPGRYQNSI GQRVCKPCDM GTYSRYGGEL CTKCPAGTVA SKTGSSQCTP CAAGFYANAP D SATSCRAC PRGYYGPYSG AYADNLGDEF EGPRGCYKCP YDFFADRPGV RQCTACPPLD LGGGNLVEQC TEDLGSQRCK PC SLLSKPK TARTEQSPPP PSPSPPPPPP PSPRPPSPNP PSPRPPSPAP PSPNPPPTSP PPSPPPSPPP PRPPPPPPPP PSP PPPNRS PPPPPPASSA INPGGGVNQN GDPVGHRRAI LSLMEDEDAE AAQEEQAIVD VDAEMQPQDD E UniProtKB: Mastigoneme-like protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, 1 mM PMSF, pH 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: -0.9 µm |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)