+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
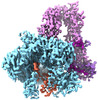
| Title | Cryo-EM structure of S. cerevisiae PolE-Ctf18-8-1-DNA | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Pol2 / Ctf18-8-1 / DNA / DNA BINDING PROTEIN-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of mitotic sister chromatid cohesion / gene conversion / DNA replication initiation / epsilon DNA polymerase complex / telomere tethering at nuclear periphery / Ctf18 RFC-like complex / maintenance of DNA trinucleotide repeats / nucleotide-excision repair, DNA gap filling / SUMO binding / Activation of the pre-replicative complex ...maintenance of mitotic sister chromatid cohesion / gene conversion / DNA replication initiation / epsilon DNA polymerase complex / telomere tethering at nuclear periphery / Ctf18 RFC-like complex / maintenance of DNA trinucleotide repeats / nucleotide-excision repair, DNA gap filling / SUMO binding / Activation of the pre-replicative complex / DNA replication proofreading / : / single-stranded DNA 3'-5' DNA exonuclease activity / mitotic DNA replication checkpoint signaling / mitotic intra-S DNA damage checkpoint signaling / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / mitotic sister chromatid cohesion / leading strand elongation / nuclear replication fork / Dual incision in TC-NER / chromosome, centromeric region / DNA replication initiation / error-prone translesion synthesis / base-excision repair, gap-filling / replication fork / double-strand break repair via homologous recombination / base-excision repair / double-strand break repair via nonhomologous end joining / DNA-templated DNA replication / double-strand break repair / mitotic cell cycle / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / double-stranded DNA binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / mRNA binding / chromatin / ATP hydrolysis activity / mitochondrion / DNA binding / zinc ion binding / ATP binding / nucleus Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Yuan Z / Georgescu R / O'Donnell M / Li H | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis. Authors: Zuanning Yuan / Roxana Georgescu / Nina Y Yao / Olga Yurieva / Michael E O'Donnell / Huilin Li /  Abstract: The proliferating cell nuclear antigen (PCNA) clamp encircles DNA to hold DNA polymerases (Pols) to DNA for processivity. The Ctf18-RFC PCNA loader, a replication factor C (RFC) variant, is specific ...The proliferating cell nuclear antigen (PCNA) clamp encircles DNA to hold DNA polymerases (Pols) to DNA for processivity. The Ctf18-RFC PCNA loader, a replication factor C (RFC) variant, is specific to the leading-strand Pol (Polε). We reveal here the underlying mechanism of Ctf18-RFC specificity to Polε using cryo-electron microscopy and biochemical studies. We found that both Ctf18-RFC and Polε contain specific structural features that direct PCNA loading onto DNA. Unlike other clamp loaders, Ctf18-RFC has a disordered ATPase associated with a diverse cellular activities (AAA+) motor that requires Polε to bind and stabilize it for efficient PCNA loading. In addition, Ctf18-RFC can pry prebound Polε off of DNA, then load PCNA onto DNA and transfer the PCNA-DNA back to Polε. These elements in both Ctf18-RFC and Polε provide specificity in loading PCNA onto DNA for Polε. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41663.map.gz emd_41663.map.gz | 10.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41663-v30.xml emd-41663-v30.xml emd-41663.xml emd-41663.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41663.png emd_41663.png | 199.7 KB | ||
| Filedesc metadata |  emd-41663.cif.gz emd-41663.cif.gz | 7.8 KB | ||
| Others |  emd_41663_half_map_1.map.gz emd_41663_half_map_1.map.gz emd_41663_half_map_2.map.gz emd_41663_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41663 http://ftp.pdbj.org/pub/emdb/structures/EMD-41663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41663 | HTTPS FTP |
-Related structure data
| Related structure data |  8tw9M  9b8rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41663.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41663.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
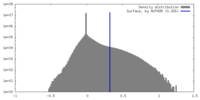
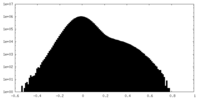
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41663_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
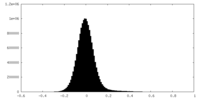
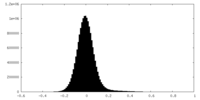
| Density Histograms |
-Half map: #1
| File | emd_41663_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
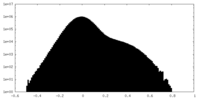
| Density Histograms |
- Sample components
Sample components
-Entire : Ctf18-RFC-PCNA complex
| Entire | Name: Ctf18-RFC-PCNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Ctf18-RFC-PCNA complex
| Supramolecule | Name: Ctf18-RFC-PCNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Chromosome transmission fidelity protein 18
| Macromolecule | Name: Chromosome transmission fidelity protein 18 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.171607 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TVKIWVKYNE GFSNAVRKNV TWNNLW UniProtKB: Chromosome transmission fidelity protein 18 |
-Macromolecule #4: DNA polymerase epsilon catalytic subunit A
| Macromolecule | Name: DNA polymerase epsilon catalytic subunit A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 255.992484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMFGKKKNNG GSSTARYSAG NKYNTLSNNY ALSAQQLLNA SKIDDIDSMM GFERYVPPQY NGRFDAKDID QIPGRVGWLT NMHATLVSQ ETLSSGSNGG GNSNDGERVT TNQGISGVDF YFLDEEGGSF KSTVVYDPYF FIACNDESRV NDVEELVKKY L ESCLKSLQ ...String: MMFGKKKNNG GSSTARYSAG NKYNTLSNNY ALSAQQLLNA SKIDDIDSMM GFERYVPPQY NGRFDAKDID QIPGRVGWLT NMHATLVSQ ETLSSGSNGG GNSNDGERVT TNQGISGVDF YFLDEEGGSF KSTVVYDPYF FIACNDESRV NDVEELVKKY L ESCLKSLQ IIRKEDLTMD NHLLGLQKTL IKLSFVNSNQ LFEARKLLRP ILQDNANNNV QRNIYNVAAN GSEKVDAKHL IE DIREYDV PYHVRVSIDK DIRVGKWYKV TQQGFIEDTR KIAFADPVVM AFDIETTKPP LKFPDSAVDQ IMMISYMIDG EGF LITNRE IISEDIEDFE YTPKPEYPGF FTIFNENDEV ALLQRFFEHI RDVRPTVIST FNGDFFDWPF IHNRSKIHGL DMFD EIGFA PDAEGEYKSS YCSHMDCFRW VKRDSYLPQG SQGLKAVTQS KLGYNPIELD PELMTPYAFE KPQHLSEYSV SDAVA TYYL YMKYVHPFIF SLCTIIPLNP DETLRKGTGT LCEMLLMVQA YQHNILLPNK HTDPIERFYD GHLLESETYV GGHVES LEA GVFRSDLKNE FKIDPSAIDE LLQELPEALK FSVEVENKSS VDKVTNFEEI KNQITQKLLE LKENNIRNEL PLIYHVD VA SMYPNIMTTN RLQPDSIKAE RDCASCDFNR PGKTCARKLK WAWRGEFFPS KMDEYNMIKR ALQNETFPNK NKFSKKKV L TFDELSYADQ VIHIKKRLTE YSRKVYHRVK VSEIVEREAI VCQRENPFYV DTVKSFRDRR YEFKGLAKTW KGNLSKIDP SDKHARDEAK KMIVLYDSLQ LAHKVILNSF YGYVMRKGSR WYSMEMAGIT CLTGATIIQM ARALVERVGR PLELDTDGIW CILPKSFPE TYFFTLENGK KLYLSYPCSM LNYRVHQKFT NHQYQELKDP LNYIYETHSE NTIFFEVDGP YKAMILPSSK E EGKGIKKR YAVFNEDGSL AELKGFELKR RGELQLIKNF QSDIFKVFLE GDTLEGCYSA VASVCNRWLD VLDSHGLMLE DE DLVSLIC ENRSMSKTLK EYEGQKSTSI TTARRLGDFL GEDMVKDKGL QCKYIISSKP FNAPVTERAI PVAIFSADIP IKR SFLRRW TLDPSLEDLD IRTIIDWGYY RERLGSAIQK IITIPAALQG VSNPVPRVEH PDWLKRKIAT KEDKFKQTSL TKFF SKTKN VPTMGKIKDI EDLFEPTVEE DNAKIKIART TKKKAVSKRK RNQLTNEEDP LVLPSEIPSM DEDYVGWLNY QKIKW KIQA RDRKRRDQLF GNTNSSRERS ALGSMIRKQA ESYANSTWEV LQYKDSGEPG VLEVFVTING KVQNITFHIP KTIYMK FKS QTMPLQKIKN CLIEKSSASL PNNPKTSNPA GGQLFKITLP ESVFLEEKEN CTSIFNDENV LGVFEGTITP HQRAIMD LG ASVTFRSKAM GALGKGIQQG FEMKDLSMAE NERYLSGFSM DIGYLLHFPT SIGYEFFSLF KSWGDTITIL VLKPSNQA Q EINASSLGQI YKQMFEKKKG KIETYSYLVD IKEDINFEFV YFTDISKLYR RLSQETTKLK EERGLQFLLL LQSPFITKL LGTIRLLNQM PIVKLSLNEV LLPQLNWQPT LLKKLVNHVL SSGSWISHLI KLSQYSNIPI CNLRLDSMDY IIDVLYARKL KKENIVLWW NEKAPLPDHG GIQNDFDLNT SWIMNDSEFP KINNSGVYDN VVLDVGVDNL TVNTILTSAL INDAEGSDLV N NNMGIDDK DAVINSPSEF VHDAFSNDAL NVLRGMLKEW WDEALKENST ADLLVNSLAS WVQNPNAKLF DGLLRYHVHN LT KKALLQL VNEFSALGST IVYADRNQIL IKTNKYSPEN CYAYSQYMMK AVRTNPMFSY LDLNIKRYWD LLIWMDKFNF SGL ACIEIE EKENQDYTAV SQWQLKKFLS PIYQPEFEDW MMIILDSMLK TKQSYLKLNS GTQRPTQIVN VKKQDKEDSV ENSL NGFSH LFSKPLMKRV KKLFKNQQEF ILDPQYEADY VIPVLPGSHL NVKNPLLELV KSLCHVMLLS KSTILEIRTL RKELL KIFE LREFAKVAEF KDPSLSLVVP DFLCEYCFFI SDIDFCKAAP ESIFSCVRCH KAFNQVLLQE HLIQKLRSDI ESYLIQ DLR CSRCHKVKRD YMSAHCPCAG AWEGTLPRES IVQKLNVFKQ VAKYYGFDIL LSCIADLTI UniProtKB: DNA polymerase epsilon catalytic subunit A |
-Macromolecule #5: Chromosome transmission fidelity protein 8
| Macromolecule | Name: Chromosome transmission fidelity protein 8 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.058493 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PSVDIDASQW QKLTQSREKQ TTVITPLGMM MLEIQGELEL PKDFASLARR DSPNEGRFSE QDGETLIRFG SLQIDGERAT LFVGKKQRL LGKVTKLDVP MGIMHFNSKD NKVELVDVMK YKVIFKDRPL PIM UniProtKB: Chromosome transmission fidelity protein 8 |
-Macromolecule #6: Sister chromatid cohesion protein DCC1
| Macromolecule | Name: Sister chromatid cohesion protein DCC1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.133785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSINLHSAPE YDPSYKLIQL TPELLDIIQD PVQNHQLRFK SLDKDKSEVV LCSHDKTWVL KQRKHSNTVL LMREFVPEQP ITFDETLLF GLSKPYMDVV GFAKTESEFE TRETHGELNL NSVPIYNGEL DFSDKIMKRS STKVIGTLEE LLENSPCSAL E GISKWHKI ...String: MSINLHSAPE YDPSYKLIQL TPELLDIIQD PVQNHQLRFK SLDKDKSEVV LCSHDKTWVL KQRKHSNTVL LMREFVPEQP ITFDETLLF GLSKPYMDVV GFAKTESEFE TRETHGELNL NSVPIYNGEL DFSDKIMKRS STKVIGTLEE LLENSPCSAL E GISKWHKI GGSVKDGVLC ILSQDFLFKA LHVLLMSAMA ESLDLQHLNV EDTHHAVGKD IEDEFNPYTR EIIETVLNKF AV QEQEAEN NTWRLRIPFI AQWYGIQALR KYVSGISMPI DEFLIKWKSL FPPFFPCDID IDMLRGYHFK PTDKTVQYIA KST LPMDPK ERFKVLFRLQ SQWDLEDIKP LIEELNSRGM KIDSFIMKYA RRKRLGKKTV VTSR UniProtKB: Sister chromatid cohesion protein DCC1 |
-Macromolecule #2: Primer DNA
| Macromolecule | Name: Primer DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.737795 KDa |
| Sequence | String: (DT)(DG)(DT)(DT)(DG)(DC)(DT)(DG)(DC) |
-Macromolecule #3: Template DNA
| Macromolecule | Name: Template DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.567998 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DA)(DG)(DC)(DA)(DG) (DC)(DA)(DA)(DC)(DA) |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 385806 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)