+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | TCR in nanodisc ND-I | |||||||||||||||||||||
 Map data Map data | unsharpened | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | T-cell receptor / TCR / 1G4 / nanodisc / CD3 / IMMUNE SYSTEM | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationdetection of tumor cell / regulation of lymphocyte apoptotic process / NNS virus cap methyltransferase / gamma-delta T cell receptor complex / Fc-gamma receptor III complex / T cell anergy / GDP polyribonucleotidyltransferase / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of T cell anergy / CD4-positive, alpha-beta T cell proliferation ...detection of tumor cell / regulation of lymphocyte apoptotic process / NNS virus cap methyltransferase / gamma-delta T cell receptor complex / Fc-gamma receptor III complex / T cell anergy / GDP polyribonucleotidyltransferase / positive regulation of cell-cell adhesion mediated by integrin / positive regulation of T cell anergy / CD4-positive, alpha-beta T cell proliferation / Fc-gamma receptor signaling pathway / gamma-delta T cell activation / negative thymic T cell selection / positive regulation of CD4-positive, alpha-beta T cell proliferation / positive thymic T cell selection / positive regulation of protein localization to cell surface / signal complex assembly / Nef and signal transduction / alpha-beta T cell receptor complex / T cell mediated cytotoxicity directed against tumor cell target / positive regulation of cell-matrix adhesion / smoothened signaling pathway / T cell receptor complex / Translocation of ZAP-70 to Immunological synapse / Phosphorylation of CD3 and TCR zeta chains / positive regulation of interleukin-4 production / establishment or maintenance of cell polarity / dendrite development / protein complex oligomerization / alpha-beta T cell activation / Generation of second messenger molecules / FCGR activation / immunological synapse / Co-inhibition by PD-1 / Role of phospholipids in phagocytosis / T cell receptor binding / T cell costimulation / positive regulation of calcium-mediated signaling / positive regulation of interleukin-2 production / positive regulation of T cell proliferation / FCGR3A-mediated IL10 synthesis / cell surface receptor protein tyrosine kinase signaling pathway / protein tyrosine kinase binding / cerebellum development / T cell activation / bioluminescence / FCGR3A-mediated phagocytosis / generation of precursor metabolites and energy / negative regulation of smoothened signaling pathway / apoptotic signaling pathway / clathrin-coated endocytic vesicle membrane / calcium-mediated signaling / Regulation of actin dynamics for phagocytic cup formation / SH3 domain binding / virion component / positive regulation of type II interferon production / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / cell-cell junction / transmembrane signaling receptor activity / Downstream TCR signaling / Cargo recognition for clathrin-mediated endocytosis / T cell receptor signaling pathway / protein transport / signaling receptor activity / Clathrin-mediated endocytosis / signaling receptor complex adaptor activity / cell body / protein-containing complex assembly / regulation of apoptotic process / dendritic spine / adaptive immune response / host cell cytoplasm / cell surface receptor signaling pathway / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / G protein-coupled receptor signaling pathway / protein heterodimerization activity / negative regulation of gene expression / hydrolase activity / external side of plasma membrane / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / positive regulation of gene expression / protein kinase binding / endoplasmic reticulum / Golgi apparatus / protein homodimerization activity / ATP binding / metal ion binding / identical protein binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   human respiratory syncytial virus human respiratory syncytial virus | |||||||||||||||||||||
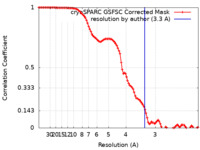
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||
 Authors Authors | Notti RQ / Walz T | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: The resting and ligand-bound states of the membrane-embedded human T-cell receptor-CD3 complex. Authors: Ryan Q Notti / Fei Yi / Søren Heissel / Martin W Bush / Zaki Molvi / Pujita Das / Henrik Molina / Christopher A Klebanoff / Thomas Walz Abstract: The T-cell receptor (TCR) initiates T-lymphocyte activation, but mechanistic questions remain( ). Here, we present cryogenic electron microscopy structures for the unliganded and human leukocyte ...The T-cell receptor (TCR) initiates T-lymphocyte activation, but mechanistic questions remain( ). Here, we present cryogenic electron microscopy structures for the unliganded and human leukocyte antigen (HLA)-bound human TCR-CD3 complex in nanodiscs that provide a native-like lipid environment. Distinct from the "open and extended" conformation seen in detergent( ), the unliganded TCR-CD3 in nanodiscs adopts two related "closed and compacted" conformations that represent its physiologic resting state . By contrast, the HLA-bound complex adopts the open and extended conformation, and conformation-locking disulfide mutants show that ectodomain opening is necessary for maximal ligand-dependent T-cell activation. Together, these results reveal allosteric conformational change during TCR activation and highlight the importance of native-like lipid environments for membrane protein structure determination. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41658.map.gz emd_41658.map.gz | 51 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41658-v30.xml emd-41658-v30.xml emd-41658.xml emd-41658.xml | 31.5 KB 31.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41658_fsc.xml emd_41658_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41658.png emd_41658.png | 34.6 KB | ||
| Filedesc metadata |  emd-41658.cif.gz emd-41658.cif.gz | 7.9 KB | ||
| Others |  emd_41658_additional_1.map.gz emd_41658_additional_1.map.gz emd_41658_additional_2.map.gz emd_41658_additional_2.map.gz emd_41658_additional_3.map.gz emd_41658_additional_3.map.gz emd_41658_additional_4.map.gz emd_41658_additional_4.map.gz emd_41658_half_map_1.map.gz emd_41658_half_map_1.map.gz emd_41658_half_map_2.map.gz emd_41658_half_map_2.map.gz | 91.5 MB 97.1 MB 97.1 MB 50.7 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41658 http://ftp.pdbj.org/pub/emdb/structures/EMD-41658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41658 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41658 | HTTPS FTP |
-Related structure data
| Related structure data |  8tw4MC  8tw6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41658.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41658.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
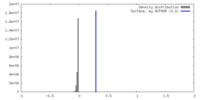
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened
| File | emd_41658_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
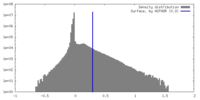
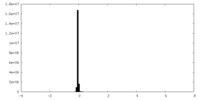
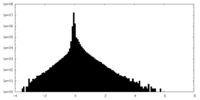
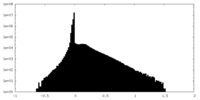
| Density Histograms |
-Additional map: unsharpened
| File | emd_41658_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
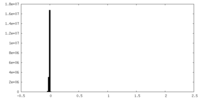
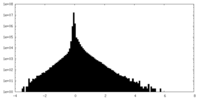
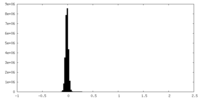
| Density Histograms |
-Additional map: #2
| File | emd_41658_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
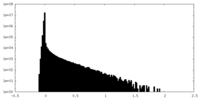
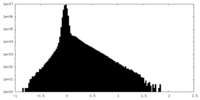
| Density Histograms |
-Additional map: #1
| File | emd_41658_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
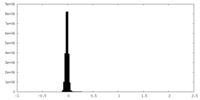
| Density Histograms |
-Half map: unsharpened
| File | emd_41658_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
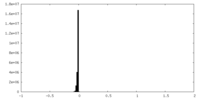
| Density Histograms |
-Half map: unsharpened
| File | emd_41658_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hetero-octameric TCR complex embedded in a lipid nanodisc
| Entire | Name: Hetero-octameric TCR complex embedded in a lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Hetero-octameric TCR complex embedded in a lipid nanodisc
| Supramolecule | Name: Hetero-octameric TCR complex embedded in a lipid nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: TCR alpha
| Macromolecule | Name: TCR alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.388189 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METLLGLLIL WLQLQWVSSK QEVTQIPAAL SVPEGENLVL NCSFTDSAIY NLQWFRQDPG KGLTSLLLIQ SSQREQTSGR LNASLDKSS GRSTLYIAAS QPGDSATYLC AVRPLYGGSY IPTFGRGTSL IVHPYIQNPD PAVYQLRDSK SSDKSVCLFT D FDSQTNVS ...String: METLLGLLIL WLQLQWVSSK QEVTQIPAAL SVPEGENLVL NCSFTDSAIY NLQWFRQDPG KGLTSLLLIQ SSQREQTSGR LNASLDKSS GRSTLYIAAS QPGDSATYLC AVRPLYGGSY IPTFGRGTSL IVHPYIQNPD PAVYQLRDSK SSDKSVCLFT D FDSQTNVS QSKDSDVYIT DKTVLDMRSM DFKSNSAVAW SNKSDFACAN AFNNSIIPED TFFPSPESSC DVKLVEKSFE TD TNLNFQN LSVIGFRILL LKVAGFNLLM TLRLWSS |
-Macromolecule #2: T cell receptor beta variable 6-5, T cell receptor beta chain MC....
| Macromolecule | Name: T cell receptor beta variable 6-5, T cell receptor beta chain MC.7.G5, MCHERRY fusion protein type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.042883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSIGLLCCAA LSLLWAGPVN AGVTQTPKFQ VLKTGQSMTL QCAQDMNHEY MSWYRQDPGM GLRLIHYSVG AGITDQGEVP NGYNVSRST TEDFPLRLLS AAPSQTSVYF CASSYVGNTG ELFFGEGSRL TVLEDLKNVF PPEVAVFEPS EAEISHTQKA T LVCLATGF ...String: MSIGLLCCAA LSLLWAGPVN AGVTQTPKFQ VLKTGQSMTL QCAQDMNHEY MSWYRQDPGM GLRLIHYSVG AGITDQGEVP NGYNVSRST TEDFPLRLLS AAPSQTSVYF CASSYVGNTG ELFFGEGSRL TVLEDLKNVF PPEVAVFEPS EAEISHTQKA T LVCLATGF YPDHVELSWW VNGKEVHSGV STDPQPLKEQ PALNDSRYCL SSRLRVSATF WQNPRNHFRC QVQFYGLSEN DE WTQDRAK PVTQIVSAEA WGRADCGFTS ESYQQGVLSA TILYEILLGK ATLYAVLVSA LVLMAMVKRK DSRGASLEVL FQG PVSKGE EDNMAIIKEF MRFKVHMEGS VNGHEFEIEG EGEGRPYEGT QTAKLKVTKG GPLPFAWDIL SPQFMYGSKA YVKH PADIP DYLKLSFPEG FKWERVMNFE DGGVVTVTQD SSLQDGEFIY KVKLRGTNFP SDGPVMQKKT MGWEASSERM YPEDG ALKG EIKQRLKLKD GGHYDAEVKT TYKAKKPVQL PGAYNVNIKL DITSHNEDYT IVEQYERAEG RHSTGGMDEL YK UniProtKB: T cell receptor beta variable 6-5, T cell receptor beta chain MC.7.G5, MCHERRY |
-Macromolecule #3: T-cell surface glycoprotein CD3 epsilon chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 epsilon chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.174227 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQSGTHWRVL GLCLLSVGVW GQDGNEEMGG ITQTPYKVSI SGTTVILTCP QYPGSEILWQ HNDKNIGGDE DDKNIGSDED HLSLKEFSE LEQSGYYVCY PRGSKPEDAN FYLYLRARVC ENCMEMDVMS VATIVIVDIC ITGGLLLLVY YWSKNRKAKA K PVTRGAGA ...String: MQSGTHWRVL GLCLLSVGVW GQDGNEEMGG ITQTPYKVSI SGTTVILTCP QYPGSEILWQ HNDKNIGGDE DDKNIGSDED HLSLKEFSE LEQSGYYVCY PRGSKPEDAN FYLYLRARVC ENCMEMDVMS VATIVIVDIC ITGGLLLLVY YWSKNRKAKA K PVTRGAGA GGRQRGQNKE RPPPVPNPDY EPIRKGQRDL YSGLNQRRI UniProtKB: T-cell surface glycoprotein CD3 epsilon chain |
-Macromolecule #4: T-cell surface glycoprotein CD3 delta chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 delta chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.949537 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEHSTFLSGL VLATLLSQVS PFKIPIEELE DRVFVNCNTS ITWVEGTVGT LLSDITRLDL GKRILDPRGI YRCNGTDIYK DKESTVQVH YRMCQSCVEL DPATVAGIIV TDVIATLLLA LGVFCFAGHE TGRLSGAADT QALLRNDQVY QPLRDRDDAQ Y SHLGGNWA RNK UniProtKB: T-cell surface glycoprotein CD3 delta chain |
-Macromolecule #5: T-cell surface glycoprotein CD3 gamma chain
| Macromolecule | Name: T-cell surface glycoprotein CD3 gamma chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.598633 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEQGKGLAVL ILAIILLQGT LAQSIKGNHL VKVYDYQEDG SVLLTCDAEA KNITWFKDGK MIGFLTEDKK KWNLGSNAKD PRGMYQCKG SQNKSKPLQV YYRMCQNCIE LNAATISGFL FAEIVSIFVL AVGVYFIAGQ DGVRQSRASD KQTLLPNDQL Y QPLKDRED DQYSHLQGNQ LRRNHHHHHH HH UniProtKB: T-cell surface glycoprotein CD3 gamma chain |
-Macromolecule #6: T-cell surface glycoprotein CD3 zeta chain,RNA-directed RNA polym...
| Macromolecule | Name: T-cell surface glycoprotein CD3 zeta chain,RNA-directed RNA polymerase L type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO / EC number: NNS virus cap methyltransferase |
|---|---|
| Source (natural) | Organism:  human respiratory syncytial virus human respiratory syncytial virus |
| Molecular weight | Theoretical: 46.650879 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKWKALFTAA ILQAQLPITE AQSFGLLDPK LCYLLDGILF IYGVILTALF LRVKFSRSAD APAYQQGQNQ LYNELNLGRR EEYDVLDKR RGRDPEMGGK PQRRKNPQEG LYNELQKDKM AEAYSEIGMK GERRRGKGHD GLYQGLSTAT KDTYDALHMQ A LPPRASLE ...String: MKWKALFTAA ILQAQLPITE AQSFGLLDPK LCYLLDGILF IYGVILTALF LRVKFSRSAD APAYQQGQNQ LYNELNLGRR EEYDVLDKR RGRDPEMGGK PQRRKNPQEG LYNELQKDKM AEAYSEIGMK GERRRGKGHD GLYQGLSTAT KDTYDALHMQ A LPPRASLE VLFQGPVSKG EELFTGVVPI LVELDGDVNG HKFSVSGEGE GDATYGKLTL KFICTTGKLP VPWPTLVTTL TY GVQCFSR YPDHMKQHDF FKSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED GNILGHKLEY NYN SHNVYI MADKQKNGIK VNFKIRHNIE DGSVQLADHY QQNTPIGDGP VLLPDNHYLS TQSKLSKDPN EKRDHMVLLE FVTA AGITL GMDELYK UniProtKB: T-cell surface glycoprotein CD3 zeta chain, Replicase |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #10: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 10 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.330 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 64000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)