[English] 日本語
 Yorodumi
Yorodumi- EMDB-41505: Hemagglutinin-neuraminidase from Human parainfluenza virus type 3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hemagglutinin-neuraminidase from Human parainfluenza virus type 3: complex with rPIV3-23 and rPIV3-28 Fabs | |||||||||
 Map data Map data | DeepEMhancer output map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Immune system / viral neutralization / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationexo-alpha-sialidase / exo-alpha-sialidase activity / host cell surface receptor binding / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Human respirovirus 3 / Human respirovirus 3 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Otrelo-Cardoso AR / Jardetzky TS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Functional and structural basis of human parainfluenza virus type 3 neutralization with human monoclonal antibodies. Authors: Naveenchandra Suryadevara / Ana Rita Otrelo-Cardoso / Nurgun Kose / Yao-Xiong Hu / Elad Binshtein / Rachael M Wolters / Alexander L Greninger / Laura S Handal / Robert H Carnahan / Anne ...Authors: Naveenchandra Suryadevara / Ana Rita Otrelo-Cardoso / Nurgun Kose / Yao-Xiong Hu / Elad Binshtein / Rachael M Wolters / Alexander L Greninger / Laura S Handal / Robert H Carnahan / Anne Moscona / Theodore S Jardetzky / James E Crowe /  Abstract: Human parainfluenza virus type 3 (hPIV3) is a respiratory pathogen that can cause severe disease in older people and infants. Currently, vaccines against hPIV3 are in clinical trials but none have ...Human parainfluenza virus type 3 (hPIV3) is a respiratory pathogen that can cause severe disease in older people and infants. Currently, vaccines against hPIV3 are in clinical trials but none have been approved yet. The haemagglutinin-neuraminidase (HN) and fusion (F) surface glycoproteins of hPIV3 are major antigenic determinants. Here we describe naturally occurring potently neutralizing human antibodies directed against both surface glycoproteins of hPIV3. We isolated seven neutralizing HN-reactive antibodies and a pre-fusion conformation F-reactive antibody from human memory B cells. One HN-binding monoclonal antibody (mAb), designated PIV3-23, exhibited functional attributes including haemagglutination and neuraminidase inhibition. We also delineated the structural basis of neutralization for two HN and one F mAbs. MAbs that neutralized hPIV3 in vitro protected against infection and disease in vivo in a cotton rat model of hPIV3 infection, suggesting correlates of protection for hPIV3 and the potential clinical utility of these mAbs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41505.map.gz emd_41505.map.gz | 51.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41505-v30.xml emd-41505-v30.xml emd-41505.xml emd-41505.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41505_fsc.xml emd_41505_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41505.png emd_41505.png | 594.6 KB | ||
| Filedesc metadata |  emd-41505.cif.gz emd-41505.cif.gz | 7.2 KB | ||
| Others |  emd_41505_half_map_1.map.gz emd_41505_half_map_1.map.gz emd_41505_half_map_2.map.gz emd_41505_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41505 http://ftp.pdbj.org/pub/emdb/structures/EMD-41505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41505 | HTTPS FTP |
-Related structure data
| Related structure data |  8tqiMC  8tqkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41505.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41505.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer output map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_41505_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
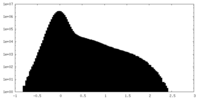
| Projections & Slices |
| ||||||||||||
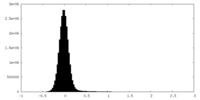
| Density Histograms |
-Half map: #1
| File | emd_41505_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
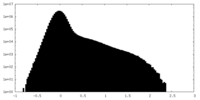
| Projections & Slices |
| ||||||||||||
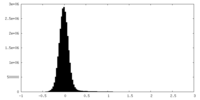
| Density Histograms |
- Sample components
Sample components
-Entire : HPIV3 HN in complex with rPIV3-23 and rPIV3-28 Fabs
| Entire | Name: HPIV3 HN in complex with rPIV3-23 and rPIV3-28 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: HPIV3 HN in complex with rPIV3-23 and rPIV3-28 Fabs
| Supramolecule | Name: HPIV3 HN in complex with rPIV3-23 and rPIV3-28 Fabs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
-Supramolecule #2: Hemagglutinin-neuraminidase glycoprotein
| Supramolecule | Name: Hemagglutinin-neuraminidase glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
-Supramolecule #3: Recombinant rPIV3-28 Fab
| Supramolecule | Name: Recombinant rPIV3-28 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Recombinant rPIV3-23 Fab
| Supramolecule | Name: Recombinant rPIV3-23 Fab / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin-neuraminidase
| Macromolecule | Name: Hemagglutinin-neuraminidase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respirovirus 3 Human respirovirus 3 |
| Molecular weight | Theoretical: 50.774414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVPPQRITHD VGIKPLNPDD FWRCTSGLPS LMKTPKIRLM PGPGLLAMPT TVDGCVRTPS LVINDLIYAY TSNLITRGCQ DIGKSYQVL QIGIITVNSD LVPDLNPRIS HTFNINDNRK SCSLALLNTD VYQLCSTPKV DERSDYASSG IEDIVLDIVN H DGSISTTR ...String: EVPPQRITHD VGIKPLNPDD FWRCTSGLPS LMKTPKIRLM PGPGLLAMPT TVDGCVRTPS LVINDLIYAY TSNLITRGCQ DIGKSYQVL QIGIITVNSD LVPDLNPRIS HTFNINDNRK SCSLALLNTD VYQLCSTPKV DERSDYASSG IEDIVLDIVN H DGSISTTR FKNNNISFDQ PYAALYPSVG PGIYYKGKII FLGYGGLEHP INENAICNTT GCPGKTQRDC NQASHSPWFS DR RMVNSII VVDKGLNSIP KLKVWTISMR QNYWGSEGRL LLLGNKIYIY TRSTSWHSKL QLGIIDITDY SDIRIKWTWH NVL SRPGNN ECPWGHSCPD GCITGVYTDA YPLNPTGSIV SSVILDSQKS RVNPVITYST STERVNELAI RNKTLSAGYT TTSC ITHYN KGYCFHIVEI NHKSLDTFQP MLFKTEIPKS CSGSENLYFQ GGSHHHHHH UniProtKB: Hemagglutinin-neuraminidase |
-Macromolecule #2: Heavy chain Fab rPIV3-28
| Macromolecule | Name: Heavy chain Fab rPIV3-28 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.344152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVKLLESGGG LIQPGDSLRL SCAASGFTFS TFAMSWVRQA PGKGLEWVSV ITSTGSSADY ADSVKGRFTM SRDNSKNTVY LQMDSLRAD DTAVYFCAKQ GATILSSFES WGQGSLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS ...String: EVKLLESGGG LIQPGDSLRL SCAASGFTFS TFAMSWVRQA PGKGLEWVSV ITSTGSSADY ADSVKGRFTM SRDNSKNTVY LQMDSLRAD DTAVYFCAKQ GATILSSFES WGQGSLVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV KDYFPEPVTV S WNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV EPKSC |
-Macromolecule #3: Light chain Fab rPIV3-28
| Macromolecule | Name: Light chain Fab rPIV3-28 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.592119 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASIGDRVT ITCQASQDID KYLNWYQQKP GKAPKLLIYD ASNFETGVPS RFSGSGSGTY FTFTISSLQA EDIATYYCQ QYDDLPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASIGDRVT ITCQASQDID KYLNWYQQKP GKAPKLLIYD ASNFETGVPS RFSGSGSGTY FTFTISSLQA EDIATYYCQ QYDDLPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLRSPVTKS FNRGEC |
-Macromolecule #4: Heavy chain Fab rPIV3-23
| Macromolecule | Name: Heavy chain Fab rPIV3-23 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.95298 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGPG LVRPSQTLSL TCSVSGASIT DDLNRWSWIR QHPGKGLECV GYISYSGTTY YNPSLQGRLS ISLDTSRNQV SLKLTSVTA ADTAVYFCAR APIIMSSPSW GLYDQDYVPM DVWGQGTTVF VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: QVQLQESGPG LVRPSQTLSL TCSVSGASIT DDLNRWSWIR QHPGKGLECV GYISYSGTTY YNPSLQGRLS ISLDTSRNQV SLKLTSVTA ADTAVYFCAR APIIMSSPSW GLYDQDYVPM DVWGQGTTVF VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSC |
-Macromolecule #5: Light chain Fab rPIV3-23
| Macromolecule | Name: Light chain Fab rPIV3-23 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.309943 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSPAT LSVSPGERVT LSCRASQSVG SDLAWYQQKP GQAPRLLIYG ASTRATGVPA KFSGSGSGTE FTLTISGLQS EDFALYYCH QYNNWWTFGL GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK ...String: DIVMTQSPAT LSVSPGERVT LSCRASQSVG SDLAWYQQKP GQAPRLLIYG ASTRATGVPA KFSGSGSGTE FTLTISGLQS EDFALYYCH QYNNWWTFGL GTKVEIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ E SVTEQDSK DSTYSLSSTL TLSKADYEKH KVYACEVTHQ GLRSPVTKSF NRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)