[English] 日本語
 Yorodumi
Yorodumi- EMDB-41285: Double nuclear outer ring of Nup84-complexes from the yeast NPC -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Double nuclear outer ring of Nup84-complexes from the yeast NPC | |||||||||||||||
 Map data Map data | full c8 ring of double Y-complexes from multibody volume | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nuclear pore complex / nucleocytoplasmic transport / nucleoporin / membrane protein / translocase / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmRNA export from nucleus in response to heat stress / positive regulation of ER to Golgi vesicle-mediated transport / Seh1-associated complex / nuclear pore localization / COPII-coated vesicle budding / protein exit from endoplasmic reticulum / nuclear pore central transport channel / COPII-mediated vesicle transport / regulation of TORC1 signaling / telomere tethering at nuclear periphery ...mRNA export from nucleus in response to heat stress / positive regulation of ER to Golgi vesicle-mediated transport / Seh1-associated complex / nuclear pore localization / COPII-coated vesicle budding / protein exit from endoplasmic reticulum / nuclear pore central transport channel / COPII-mediated vesicle transport / regulation of TORC1 signaling / telomere tethering at nuclear periphery / nuclear pore outer ring / positive regulation of protein exit from endoplasmic reticulum / : / COPII vesicle coat / Regulation of HSF1-mediated heat shock response / post-transcriptional tethering of RNA polymerase II gene DNA at nuclear periphery / SUMOylation of SUMOylation proteins / structural constituent of nuclear pore / SUMOylation of RNA binding proteins / SUMOylation of chromatin organization proteins / silent mating-type cassette heterochromatin formation / vacuolar membrane / nucleocytoplasmic transport / ribosomal large subunit export from nucleus / positive regulation of TOR signaling / mRNA transport / nuclear pore / subtelomeric heterochromatin formation / mRNA export from nucleus / ERAD pathway / positive regulation of TORC1 signaling / protein export from nucleus / cell periphery / protein import into nucleus / nuclear envelope / double-strand break repair / nuclear membrane / chromosome, telomeric region / endoplasmic reticulum membrane / positive regulation of DNA-templated transcription / structural molecule activity / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / positive regulation of transcription by RNA polymerase II / identical protein binding Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
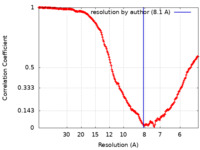
| Method | single particle reconstruction / cryo EM / Resolution: 8.1 Å | |||||||||||||||
 Authors Authors | Akey CW / Echeverria I / Ouch C / Fernandez-Martinez J / Rout MP | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Implications of a multiscale structure of the yeast nuclear pore complex. Authors: Christopher W Akey / Ignacia Echeverria / Christna Ouch / Ilona Nudelman / Yi Shi / Junjie Wang / Brian T Chait / Andrej Sali / Javier Fernandez-Martinez / Michael P Rout /   Abstract: Nuclear pore complexes (NPCs) direct the nucleocytoplasmic transport of macromolecules. Here, we provide a composite multiscale structure of the yeast NPC, based on improved 3D density maps from ...Nuclear pore complexes (NPCs) direct the nucleocytoplasmic transport of macromolecules. Here, we provide a composite multiscale structure of the yeast NPC, based on improved 3D density maps from cryogenic electron microscopy and AlphaFold2 models. Key features of the inner and outer rings were integrated into a comprehensive model. We resolved flexible connectors that tie together the core scaffold, along with equatorial transmembrane complexes and a lumenal ring that anchor this channel within the pore membrane. The organization of the nuclear double outer ring reveals an architecture that may be shared with ancestral NPCs. Additional connections between the core scaffold and the central transporter suggest that under certain conditions, a degree of local organization is present at the periphery of the transport machinery. These connectors may couple conformational changes in the scaffold to the central transporter to modulate transport. Collectively, this analysis provides insights into assembly, transport, and NPC evolution. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41285.map.gz emd_41285.map.gz | 9.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41285-v30.xml emd-41285-v30.xml emd-41285.xml emd-41285.xml | 43 KB 43 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41285_fsc.xml emd_41285_fsc.xml | 21.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_41285.png emd_41285.png | 160.3 KB | ||
| Filedesc metadata |  emd-41285.cif.gz emd-41285.cif.gz | 11.1 KB | ||
| Others |  emd_41285_additional_1.map.gz emd_41285_additional_1.map.gz emd_41285_additional_2.map.gz emd_41285_additional_2.map.gz emd_41285_additional_3.map.gz emd_41285_additional_3.map.gz emd_41285_additional_4.map.gz emd_41285_additional_4.map.gz emd_41285_half_map_1.map.gz emd_41285_half_map_1.map.gz emd_41285_half_map_2.map.gz emd_41285_half_map_2.map.gz | 5.6 MB 1 MB 5.6 MB 96.2 MB 31.9 MB 31.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41285 http://ftp.pdbj.org/pub/emdb/structures/EMD-41285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41285 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41285 | HTTPS FTP |
-Related structure data
| Related structure data |  8tieMC  8t9lC  8tj5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41285.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41285.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full c8 ring of double Y-complexes from multibody volume | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.66 Å | ||||||||||||||||||||||||||||||||||||
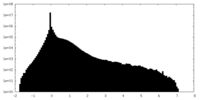
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: multibody half map
| File | emd_41285_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | multibody half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
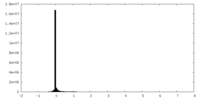
| Density Histograms |
-Additional map: recombined 2 Y-complex protomer, bfactor -600 and local...
| File | emd_41285_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | recombined 2 Y-complex protomer, bfactor -600 and local resolution box 120 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: multibody half map
| File | emd_41285_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | multibody half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
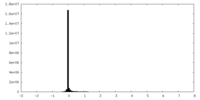
| Density Histograms |
-Additional map: post processed multibody volume
| File | emd_41285_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post processed multibody volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
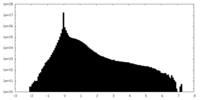
| Density Histograms |
-Half map: half map of c8 ring
| File | emd_41285_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of c8 ring | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map of c8 ring
| File | emd_41285_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of c8 ring | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nuclear Pore Complex
| Entire | Name: Nuclear Pore Complex |
|---|---|
| Components |
|
-Supramolecule #1: Nuclear Pore Complex
| Supramolecule | Name: Nuclear Pore Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Protein A tagged Mlp1 pullout of NPC |
|---|---|
| Source (natural) | Organism:  Strain: MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 MLP1-PPX-ProteinA::HIS5 Organelle: nucleus / Location in cell: nuclear envelope |
-Supramolecule #2: double Nup84 Y-complex
| Supramolecule | Name: double Nup84 Y-complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: all / Details: Nup84 Y-complexes in nuclear double outer ring |
|---|---|
| Source (natural) | Organism:  Strain: MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 MLP1-PPX-ProteinA::HIS5 Organelle: nucleus / Location in cell: nuclear envelope |
-Macromolecule #1: Nucleoporin NUP120
| Macromolecule | Name: Nucleoporin NUP120 / type: protein_or_peptide / ID: 1 / Details: long arm of Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 120.560328 KDa |
| Sequence | String: MACLSRIDAN LLQYYEKPEP NNTVDLYVSN NSNNNGLKEG DKSISTPVPQ PYGSEYSNCL LLSNSEYICY HFSSRSTLLT FYPLSDAYH GKTINIHLPN ASMNQRYTLT IQEVEQQLLV NVILKDGSFL TLQLPLSFLF SSANTLNGEW FHLQNPYDFT V RVPHFLFY ...String: MACLSRIDAN LLQYYEKPEP NNTVDLYVSN NSNNNGLKEG DKSISTPVPQ PYGSEYSNCL LLSNSEYICY HFSSRSTLLT FYPLSDAYH GKTINIHLPN ASMNQRYTLT IQEVEQQLLV NVILKDGSFL TLQLPLSFLF SSANTLNGEW FHLQNPYDFT V RVPHFLFY VSPQFSVVFL EDGGLLGLKK VDGVHYEPLL FNDNSYLKSL TRFFSRSSKS DYDSVISCKL FHERYLIVLT QN CHLKIWD LTSFTLIQDY DMVSQSDSDP SHFRKVEAVG EYLSLYNNTL VTLLPLENGL FQMGTLLVDS SGILTYTFQN NIP TNLSAS AIWSIVDLVL TRPLELNVEA SYLNLIVLWK SGTASKLQIL NVNDESFKNY EWIESVNKSL VDLQSEHDLD IVTK TGDVE RGFCNLKSRY GTQIFERAQQ ILSENKIIMA HNEDEEYLAN LETILRDVKT AFNEASSITL YGDEIILVNC FQPYN HSLY KLNTTVENWF YNMHSETDGS ELFKYLRTLN GFASTLSNDV LRSISKKFLD IITGELPDSM TTVEKFTDIF KNCLEN QFE ITNLKILFDE LNSFDIPVVL NDLINNQMKP GIFWKKDFIS AIKFDGFTSI ISLESLHQLL SIHYRITLQV LLTFVLF DL DTEIFGQHIS TLLDLHYKQF LLLNLYRQDK CLLAEVLLKD SSEFSFGVKF FNYGQLIAYI DSLNSNVYNA SITENSFF M TFFRSYIIEN TSHKNIRFFL ENVECPFYLR HNEVQEFMFA MTLFSCGNFD QSYEIFQLHD YPEAINDKLP TFLEDLKSE NYHGDSIWKD LLCTFTVPYR HSAFYYQLSL LFDRNNSQEF ALKCISKSAE YSLKEIQIEE LQDFKEKQHI HYLNLLIHFR MFEEVLDVL RLGHECLSDT VRTNFLQLLL QEDIYSRDFF STLLRLCNAH SDNGELYLRT VDIKIVDSIL SQNLRSGDWE C FKKLYCFR MLNKSERAAA EVLYQYILMQ ADLDVIRKRK CYLMVINVLS SFDSAYDQWI LNGSKVVTLT DLRDELRGL UniProtKB: Nucleoporin NUP120 |
-Macromolecule #2: Nucleoporin NUP85
| Macromolecule | Name: Nucleoporin NUP85 / type: protein_or_peptide / ID: 2 / Details: short arm of Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.972438 KDa |
| Sequence | String: MTIDDSNRLL MDVDQFDFLD DGTAQLSNNK TDEEEQLYKR DPVSGAILVP MTVNDQPIEK NGDKMPLKFK LGPLSYQNMA FITAKDKYK LYPVRIPRLD TSKEFSAYVS GLFEIYRDLG DDRVFNVPTI GVVNSNFAKE HNATVNLAME AILNELEVFI G RVKDQDGR ...String: MTIDDSNRLL MDVDQFDFLD DGTAQLSNNK TDEEEQLYKR DPVSGAILVP MTVNDQPIEK NGDKMPLKFK LGPLSYQNMA FITAKDKYK LYPVRIPRLD TSKEFSAYVS GLFEIYRDLG DDRVFNVPTI GVVNSNFAKE HNATVNLAME AILNELEVFI G RVKDQDGR VNRFYELEES LTVLNCLRTM YFILDGQDVE ENRSEFIESL LNWINRSDGE PDEEYIEQVF SVKDSTAGKK VF ETQYFWK LLNQLVLRGL LSQAIGCIER SDLLPYLSDT CAVSFDAVSD SIELLKQYPK DSSSTFREWK NLVLKLSQAF GSS ATDISG ELRDYIEDFL LVIGGNQRKI LQYSRTWYES FCGFLLYYIP SLELSAEYLQ MSLEANVVDI TNDWEQPCVD IISG KIHSI LPVMESLDSC TAAFTAMICE AKGLIENIFE GEKNSDDYSN EDNEMLEDLF SYRNGMASYM LNSFAFELCS LGDKE LWPV AIGLIALSAT GTRSAKKMVI AELLPHYPFV TNDDIEWMLS ICVEWRLPEI AKEIYTTLGN QMLSAHNIIE SIANFS RAG KYELVKSYSW LLFEASCMEG QKLDDPVLNA IVSKNSPAED DVIIPQDILD CVVTNSMRQT LAPYAVLSQF YELRDRE DW GQALRLLLLL IEFPYLPKHY LVLLVAKFLY PIFLLDDKKL MDEDSVATVI EVIETKWDDA DEKSSNLYET IIEADKSL P SSMATLLKNL RKKLNFKLCQ AFM UniProtKB: Nucleoporin NUP85 |
-Macromolecule #3: NUP145 isoform 1
| Macromolecule | Name: NUP145 isoform 1 / type: protein_or_peptide / ID: 3 / Details: stem junction of Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 145.810578 KDa |
| Sequence | String: MFNKSVNSGF TFGNQNTSTP TSTPAQPSSS LQFPQKSTGL FGNVNVNANT STPSPSGGLF NANSNANSIS QQPANNSLFG NKPAQPSGG LFGATNNTTS KSAGSLFGNN NATANSTGST GLFSGSNNIA SSTQNGGLFG NSNNNNITST TQNGGLFGKP T TTPAGAGG ...String: MFNKSVNSGF TFGNQNTSTP TSTPAQPSSS LQFPQKSTGL FGNVNVNANT STPSPSGGLF NANSNANSIS QQPANNSLFG NKPAQPSGG LFGATNNTTS KSAGSLFGNN NATANSTGST GLFSGSNNIA SSTQNGGLFG NSNNNNITST TQNGGLFGKP T TTPAGAGG LFGNSSSTNS TTGLFGSNNT QSSTGIFGQK PGASTTGGLF GNNGASFPRS GETTGTMSTN PYGINISNVP MA VADMPRS ITSSLSDVNG KSDAEPKPIE NRRTYSFSSS VSGNAPLPLA SQSSLVSRLS TRLKATQKST SPNEIFSPSY SKP WLNGAG SAPLVDDFFS SKMTSLAPNE NSIFPQNGFN FLSSQRADLT ELRKLKIDSN RSAAKKLKLL SGTPAITKKH MQDE QDSSE NEPIANADSV TNIDRKENRD NNLDNTYLNG KEQSNNLNKQ DGENTLQHEK SSSFGYWCSP SPEQLERLSL KQLAA VSNF VIGRRGYGCI TFQHDVDLTA FTKSFREELF GKIVIFRSSK TVEVYPDEAT KPMIGHGLNV PAIITLENVY PVDKKT KKP MKDTTKFAEF QVFDRKLRSM REMNYISYNP FGGTWTFKVN HFSIWGLVNE EDAEIDEDDL SKQEDGGEQP LRKVRTL AQ SKPSDKEVIL KTDGTFGTLS GKDDSIVEEK AYEPDLSDAD FEGIEASPKL DVSKDWVEQL ILAGSSLRSV FATSKEFD G PCQNEIDLLF SECNDEIDNA KLIMKERRFT ASYTFAKFST GSMLLTKDIV GKSGVSIKRL PTELQRKFLF DDVYLDKEI EKVTIEARKS NPYPQISESS LLFKDALDYM EKTSSDYNLW KLSSILFDPV SYPYKTDNDQ VKMALLKKER HCRLTSWIVS QIGPEIEEK IRNSSNEIEQ IFLYLLLNDV VRASKLAIES KNGHLSVLIS YLGSNDPRIR DLAELQLQKW STGGCSIDKN I SKIYKLLS GSPFEGLFSL KELESEFSWL CLLNLTLCYG QIDEYSLESL VQSHLDKFSL PYDDPIGVIF QLYAANENTE KL YKEVRQR TNALDVQFCW YLIQTLRFNG TRVFSKETSD EATFAFAAQL EFAQLHGHSL FVSCFLNDDK AAEDTIKRLV MRE ITLLRA STNDHILNRL KIPSQLIFNA QALKDRYEGN YLSEVQNLLL GSSYDLAEMA IVTSLGPRLL LSNNPVQNNE LKTL REILN EFPDSERDKW SVSINVFEVY LKLVLDNVET QETIDSLISG MKIFYDQYKH CREVAACCNV MSQEIVSKIL EKNNP SIGD SKAKLLELPL GQPEKAYLRG EFAQDLMKCT YKI UniProtKB: NUP145 isoform 1 |
-Macromolecule #4: Protein transport protein SEC13
| Macromolecule | Name: Protein transport protein SEC13 / type: protein_or_peptide / ID: 4 / Details: cofactor of Nup145C on stem of Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.082965 KDa |
| Sequence | String: MVVIANAHNE LIHDAVLDYY GKRLATCSSD KTIKIFEVEG ETHKLIDTLT GHEGPVWRVD WAHPKFGTIL ASCSYDGKVL IWKEENGRW SQIAVHAVHS ASVNSVQWAP HEYGPLLLVA SSDGKVSVVE FKENGTTSPI IIDAHAIGVN SASWAPATIE E DGEHNGTK ...String: MVVIANAHNE LIHDAVLDYY GKRLATCSSD KTIKIFEVEG ETHKLIDTLT GHEGPVWRVD WAHPKFGTIL ASCSYDGKVL IWKEENGRW SQIAVHAVHS ASVNSVQWAP HEYGPLLLVA SSDGKVSVVE FKENGTTSPI IIDAHAIGVN SASWAPATIE E DGEHNGTK ESRKFVTGGA DNLVKIWKYN SDAQTYVLES TLEGHSDWVR DVAWSPTVLL RSYLASVSQD RTCIIWTQDN EQ GPWKKTL LKEEKFPDVL WRASWSLSGN VLALSGGDNK VTLWKENLEG KWEPAGEVHQ UniProtKB: Protein transport protein SEC13 |
-Macromolecule #5: Nucleoporin Seh1
| Macromolecule | Name: Nucleoporin Seh1 / type: protein_or_peptide / ID: 5 / Details: cofactor Nup85 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.652078 KDa |
| Sequence | String: MQPFDSGHDD LVHDVVYDFY GRHVATCSSD QHIKVFKLDK DTSNWELSDS WRAHDSSIVA IDWASPEYGR IIASASYDKT VKLWEEDPD QEECSGRRWN KLCTLNDSKG SLYSVKFAPA HLGLKLACLG NDGILRLYDA LEPSDLRSWT LTSEMKVLSI P PANHLQSD ...String: MQPFDSGHDD LVHDVVYDFY GRHVATCSSD QHIKVFKLDK DTSNWELSDS WRAHDSSIVA IDWASPEYGR IIASASYDKT VKLWEEDPD QEECSGRRWN KLCTLNDSKG SLYSVKFAPA HLGLKLACLG NDGILRLYDA LEPSDLRSWT LTSEMKVLSI P PANHLQSD FCLSWCPSRF SPEKLAVSAL EQAIIYQRGK DGKLHVAAKL PGHKSLIRSI SWAPSIGRWY QLIATGCKDG RI RIFKITE KNLQVELLSE HDDHNGEVWS VSWNLTGTIL SSAGDDGKVR LWKATYSNEF KCMSVITAQQ |
-Macromolecule #6: Nucleoporin NUP84
| Macromolecule | Name: Nucleoporin NUP84 / type: protein_or_peptide / ID: 6 / Details: tail of Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 83.718867 KDa |
| Sequence | String: MELSPTYQTE RFTKFSDTLK EFKIEQNNEQ NPIDPFNIIR EFRSAAGQLA LDLANSGDES NVISSKDWEL EARFWHLVEL LLVFRNADL DLDEMELHPY NSRGLFEKKL MQDNKQLYQI WIVMVWLKEN TYVMERPKNV PTSKWLNSIT SGGLKSCDLD F PLRENTNV ...String: MELSPTYQTE RFTKFSDTLK EFKIEQNNEQ NPIDPFNIIR EFRSAAGQLA LDLANSGDES NVISSKDWEL EARFWHLVEL LLVFRNADL DLDEMELHPY NSRGLFEKKL MQDNKQLYQI WIVMVWLKEN TYVMERPKNV PTSKWLNSIT SGGLKSCDLD F PLRENTNV LDVKDKEEDH IFFKYIYELI LAGAIDEALE EAKLSDNISI CMILCGIQEY LNPVIDTQIA NEFNTQQGIK KH SLWRRTV YSLSQQAGLD PYERAIYSYL SGAIPNQEVL QYSDWESDLH IHLNQILQTE IENYLLENNQ VGTDELILPL PSH ALTVQE VLNRVASRHP SESEHPIRVL MASVILDSLP SVIHSSVEML LDVVKGTEAS NDIIDKPYLL RIVTHLAICL DIIN PGSVE EVDKSKLITT YISLLKLQGL YENIPIYATF LNESDCLEAC SFILSSLEDP QVRKKQIETI NFLRLPASNI LRRTT QRVF DETEQEYSPS NEISISFDVN NIDMHLIYGV EWLIEGKLYV DAVHSIIALS RRFLLNGRVK ALEQFMERNN IGEICK NYE LEKIADNISK DENEDQFLEE ITQYEHLIKG IREYEEWQKS VSLLSSESNI PTLIEKLQGF SKDTFELIKT FLVDLTS SN FADSADYEIL YEIRALYTPF LLMELHKKLV EAAKLLKIPK FISEALAFTS LVANENDKIY LLFQSSGKLK EYLDLVAR T ATLSN UniProtKB: Nucleoporin NUP84 |
-Macromolecule #7: NUP133 isoform 1
| Macromolecule | Name: NUP133 isoform 1 / type: protein_or_peptide / ID: 7 / Details: tail Nup84 Y-complex / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.452672 KDa |
| Sequence | String: MSEKKVHLRL RKELSVPIAV VENESLAQLS YEEESQASLM DISMEQQQLR LHSHFDNSKV FTENNRYIVK TLQTDYSSGF SNDDELNGY IDMQIGYGLV NDHKKVYIWN IHSTQKDTPY ITVPFRSDDN DEIAVAPRCI LTFPATMDES PLALNPNDQD E TGGLIIIK ...String: MSEKKVHLRL RKELSVPIAV VENESLAQLS YEEESQASLM DISMEQQQLR LHSHFDNSKV FTENNRYIVK TLQTDYSSGF SNDDELNGY IDMQIGYGLV NDHKKVYIWN IHSTQKDTPY ITVPFRSDDN DEIAVAPRCI LTFPATMDES PLALNPNDQD E TGGLIIIK GSKAIYYEDI NSINNLNFKL SEKFSHELEL PINSSGGEKC DLMLNCEPAG IVLSTNMGRI FFITIRNSMG KP QLKLGKL LNKPFKLGIW SKIFNTNSSV VSLRNGPILG KGTRLVYITT NKGIFQTWQL SATNSHPTKL IDVNIYEAIL ESL QDLYPF AHGTLKIWDS HPLQDESSQL FLSSIYDSSC NETYYILSTI IFDSSSNSFT IFSTYRLNTF MESITDTKFK PKIF IPQME NANDTNEVTS ILVMFPNAVV ITQVNSKLDS SYSMRRKWED IVSLRNDIDI IGSGYDSKSL YVLTKQMGVL QFFVK ENEE TNSKPEVGFV KSHVDQAVYF SKINANPIDF NLPPEISLDQ ESIEHDLKLT SEEIFHSNGK YIPPMLNTLG QHLSVR KEF FQNFLTFVAK NFNYKISPEL KLDLIEKFEI LNCCIKFNSI IRQSDVLNDI WEKTLSNYNL TQNEHLTTKT VVINSPD VF PVIFKQFLNH VVFVLFPSQN QNFKLNVTNL INLCFYDGIL EEGEKTIRYE LLELDPMEVD TSKLPWFINF DYLNCINQ C FFDFTFACEE EGSLDSYKEG LLKIVKILYY QFNQFKIWIN TQPVKSVNAN DNFININNLY DDNHLDWNHV LCKVNLKEQ CIQIAEFYKD LSGLVQTLQT LDQNDSTTVS LYETFFNEFP KEFSFTLFEY LIKHKKLNDL IFRFPQQHDV LIQFFQESAP KYGHVAWIQ QILDGSYADA MNTLKNITVD DSKKGESLSE CELHLNVAKL SSLLVEKDNL DINTLRKIQY NLDTIDAEKN I SNKLKKGE VQICKRFKNG SIREVFNILV EELKSTTVVN LSDLVELYSM LDDEESLFIP LRLLSVDGNL LNFEVKKFLN AL VWRRIVL LNASNEGDKL LQHIVKRVFD EELPKNNDFP LPSVDLLCDK SLLTPEYISE TYGRFPIDQN AIREEIYEEI SQV ETLNSD NSLEIKLHST IGSVAKEKNY TINYETNTVE Y UniProtKB: NUP133 isoform 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20mM HEPES,50mM Potassium acetate,20mM NaCl,2mM MgCl2,1mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK III |
| Details | One step affinity purified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Preliminary grid screening done manually with individual images of low magnification montages of candidate meshes. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-40 / Number grids imaged: 1 / Number real images: 4015 / Average electron dose: 40.0 e/Å2 / Details: 3218 images retained after triage |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 37651 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: experimental model / Details: none |
|---|---|
| Details | Molecular dynamics flexible fitting after docking Alphafold2 CSMs for Nups into the 3D map of double Y complex ring with Chimera |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: cross correlation |
| Output model |  PDB-8tie: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)