[English] 日本語
 Yorodumi
Yorodumi- EMDB-41282: CryoEM structure of octamer assembly of Shedu nuclease domain fro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of octamer assembly of Shedu nuclease domain from Bacillus cereus | |||||||||
 Map data Map data | CryoEM structure of Shedu from Bacillus cereus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Shedu / DUF4263 / Bacterial defense systems / Nuclease / Anti-plasmid defense system / PD-(D/E)XK nuclease / Whirly domain / Two-component signaling / DNA BINDING PROTEIN | |||||||||
| Function / homology | Protein of unknown function DUF4263 / : / Shedu protein SduA, C-terminal / Shedu protein SduA, N-terminal / nuclease activity / defense response to virus / hydrolase activity / Shedu protein SduA Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
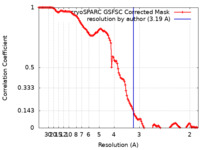
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Gu Y / Corbett K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2025 Journal: Mol Cell / Year: 2025Title: Bacterial Shedu immune nucleases share a common enzymatic core regulated by diverse sensor domains. Authors: Yajie Gu / Huan Li / Amar Deep / Eray Enustun / Dapeng Zhang / Kevin D Corbett /  Abstract: Prokaryotes possess diverse anti-bacteriophage immune systems, including the single-protein Shedu nuclease. Here, we reveal the structural basis for activation of Bacillus cereus Shedu. Two ...Prokaryotes possess diverse anti-bacteriophage immune systems, including the single-protein Shedu nuclease. Here, we reveal the structural basis for activation of Bacillus cereus Shedu. Two cryoelectron microscopy structures of Shedu show that it switches between inactive and active states through conformational changes affecting active-site architecture, which are controlled by the protein's N-terminal domain (NTD). We find that B. cereus Shedu cleaves near DNA ends with a 3' single-stranded overhang, likely enabling it to specifically degrade the DNA injected by certain bacteriophages. Bioinformatic analysis of Shedu homologs reveals a conserved nuclease domain with remarkably diverse N-terminal regulatory domains: we identify 79 distinct NTD types falling into eight broad classes, including those with predicted nucleic acid binding, enzymatic, and other activities. Together, these data reveal Shedu as a broad family of immune nucleases with a common nuclease core regulated by diverse NTDs that likely respond to a range of signals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41282.map.gz emd_41282.map.gz | 89.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41282-v30.xml emd-41282-v30.xml emd-41282.xml emd-41282.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41282_fsc.xml emd_41282_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_41282.png emd_41282.png | 94.1 KB | ||
| Filedesc metadata |  emd-41282.cif.gz emd-41282.cif.gz | 6.5 KB | ||
| Others |  emd_41282_additional_1.map.gz emd_41282_additional_1.map.gz emd_41282_additional_2.map.gz emd_41282_additional_2.map.gz emd_41282_additional_3.map.gz emd_41282_additional_3.map.gz emd_41282_half_map_1.map.gz emd_41282_half_map_1.map.gz emd_41282_half_map_2.map.gz emd_41282_half_map_2.map.gz | 89.6 MB 165.1 MB 165.1 MB 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41282 http://ftp.pdbj.org/pub/emdb/structures/EMD-41282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41282 | HTTPS FTP |
-Related structure data
| Related structure data |  8ti9MC  8tiaMC  8ti8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41282.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41282.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of Shedu from Bacillus cereus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
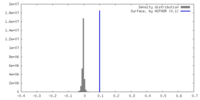
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: focused local refinement, main map
| File | emd_41282_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused local refinement, main map | ||||||||||||
| Projections & Slices |
| ||||||||||||
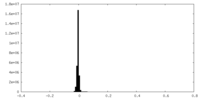
| Density Histograms |
-Additional map: focused local refinement, half A map
| File | emd_41282_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused local refinement, half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
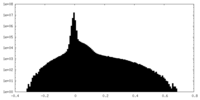
| Density Histograms |
-Additional map: focused local refinement, half B map
| File | emd_41282_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused local refinement, half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
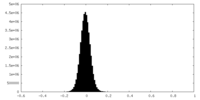
| Density Histograms |
-Half map: Half Map 1
| File | emd_41282_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_41282_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Bacillus cereus Shedu delta_NL octamer
| Entire | Name: Bacillus cereus Shedu delta_NL octamer |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus cereus Shedu delta_NL octamer
| Supramolecule | Name: Bacillus cereus Shedu delta_NL octamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Octamer assembly of Bacillus cereus Shedu nuclease domain, which truncates the N-terminal and linker region. Glutamic acid 264 is mutated to Alanine. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 216 KDa |
-Macromolecule #1: Shedu protein SduA
| Macromolecule | Name: Shedu protein SduA / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.23676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAK EQDLDQLNTL IGIANLKKVL SVWESNKLTN TSEKFWQSVL KENTWILSQI FSNPTVLIND EAYVGGKTV KNDSGKLVDF LYANPFSKDA VLIAIKTPST PLITPTEYRT GVYSAHKDLT GAVTQVLTYK TTLQREYQNI D YNNYRQGI ...String: MKSSHHHHHH ENLYFQSNAK EQDLDQLNTL IGIANLKKVL SVWESNKLTN TSEKFWQSVL KENTWILSQI FSNPTVLIND EAYVGGKTV KNDSGKLVDF LYANPFSKDA VLIAIKTPST PLITPTEYRT GVYSAHKDLT GAVTQVLTYK TTLQREYQNI D YNNYRQGI KTDFDIITPC CVVIAGMFDT LTDTAHRHSF ELYRKELKNV TVITFDELFE RVKGLIKLLE G UniProtKB: Shedu protein SduA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Prepared using deionized water and filtered strelized. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Freshly collected from size-exclusion column |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Detector mode: COUNTING / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8ti9:  PDB-8tia: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)