[English] 日本語
 Yorodumi
Yorodumi- EMDB-41251: Linker domain of Nexin-dynein regulatory complex from Tetrahymena... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Linker domain of Nexin-dynein regulatory complex from Tetrahymena thermophila | |||||||||
 Map data Map data | Primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nexin-dynein regulatory complex / cilia / axoneme / dynein / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of cilium movement / axonemal dynein complex assembly / axonemal dynein complex / cilium-dependent cell motility / cilium organization / myosin II complex / nucleocytoplasmic transport / motile cilium / cilium assembly / axoneme ...regulation of cilium movement / axonemal dynein complex assembly / axonemal dynein complex / cilium-dependent cell motility / cilium organization / myosin II complex / nucleocytoplasmic transport / motile cilium / cilium assembly / axoneme / GTPase activator activity / cell motility / small GTPase binding / spermatogenesis / microtubule binding / microtubule / cytoskeleton / cell differentiation / cilium / ciliary basal body / calcium ion binding / perinuclear region of cytoplasm / Golgi apparatus / ATP hydrolysis activity / ATP binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
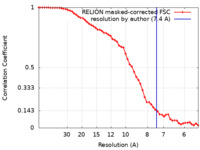
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||
 Authors Authors | Ghanaeian AG / Bui KH | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism. Authors: Avrin Ghanaeian / Sumita Majhi / Caitlyn L McCafferty / Babak Nami / Corbin S Black / Shun Kai Yang / Thibault Legal / Ophelia Papoulas / Martyna Janowska / Melissa Valente-Paterno / Edward ...Authors: Avrin Ghanaeian / Sumita Majhi / Caitlyn L McCafferty / Babak Nami / Corbin S Black / Shun Kai Yang / Thibault Legal / Ophelia Papoulas / Martyna Janowska / Melissa Valente-Paterno / Edward M Marcotte / Dorota Wloga / Khanh Huy Bui /    Abstract: Cilia are hairlike protrusions that project from the surface of eukaryotic cells and play key roles in cell signaling and motility. Ciliary motility is regulated by the conserved nexin-dynein ...Cilia are hairlike protrusions that project from the surface of eukaryotic cells and play key roles in cell signaling and motility. Ciliary motility is regulated by the conserved nexin-dynein regulatory complex (N-DRC), which links adjacent doublet microtubules and regulates and coordinates the activity of outer doublet complexes. Despite its critical role in cilia motility, the assembly and molecular basis of the regulatory mechanism are poorly understood. Here, using cryo-electron microscopy in conjunction with biochemical cross-linking and integrative modeling, we localize 12 DRC subunits in the N-DRC structure of Tetrahymena thermophila. We also find that the CCDC96/113 complex is in close contact with the DRC9/10 in the linker region. In addition, we reveal that the N-DRC is associated with a network of coiled-coil proteins that most likely mediates N-DRC regulatory activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
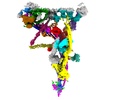
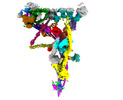
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41251.map.gz emd_41251.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41251-v30.xml emd-41251-v30.xml emd-41251.xml emd-41251.xml | 33.1 KB 33.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41251_fsc.xml emd_41251_fsc.xml | 5.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_41251.jpg emd_41251.jpg emd_41251.png emd_41251.png | 372.5 KB 70.2 KB | ||
| Masks |  emd_41251_msk_1.map emd_41251_msk_1.map | 12.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41251.cif.gz emd-41251.cif.gz | 10.7 KB | ||
| Others |  emd_41251_half_map_1.map.gz emd_41251_half_map_1.map.gz emd_41251_half_map_2.map.gz emd_41251_half_map_2.map.gz | 9.8 MB 9.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41251 http://ftp.pdbj.org/pub/emdb/structures/EMD-41251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41251 | HTTPS FTP |
-Related structure data
| Related structure data |  8th8MC  8tekC  8tidC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41251.map.gz / Format: CCP4 / Size: 43.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41251.map.gz / Format: CCP4 / Size: 43.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.74 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41251_msk_1.map emd_41251_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_41251_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_41251_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Linker domain of Nexin-dynein regulatory complex from Tetrahymena...
+Supramolecule #1: Linker domain of Nexin-dynein regulatory complex from Tetrahymena...
+Macromolecule #1: Dynein regulatory complex protein 1/2 N-terminal domain-containin...
+Macromolecule #2: Coiled-coil protein, putative
+Macromolecule #3: LRRC48 protein
+Macromolecule #4: Growth-arrest-specific microtubule-binding protein
+Macromolecule #5: Growth-arrest-specific microtubule-binding protein
+Macromolecule #6: Flagellar associated protein
+Macromolecule #7: Kinase domain protein
+Macromolecule #8: Coiled-coil lobo-like protein, putative
+Macromolecule #9: EF-hand calcium-binding domain protein
+Macromolecule #10: Dynein regulatory complex protein 9
+Macromolecule #11: Dynein regulatory complex protein 10
+Macromolecule #12: AAA family ATPase CDC48 subfamily protein
+Macromolecule #13: DUF4201 domain-containing protein
+Macromolecule #14: Calmodulin 7-2
+Macromolecule #15: Coiled-coil domain-containing protein 153
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)