[English] 日本語
 Yorodumi
Yorodumi- EMDB-41198: Tropomyosin-receptor kinase fused gene protein (TRK-fused gene pr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | amyloid / mutation / Charcot-Marie-Tooth disease / Hereditary motor and sensory neuropathy with proximal dominant involvement / PROTEIN FIBRIL | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationCOPII vesicle coating / COPII-mediated vesicle transport / endoplasmic reticulum exit site / endoplasmic reticulum to Golgi vesicle-mediated transport / bioluminescence / generation of precursor metabolites and energy / Signaling by ALK fusions and activated point mutants / positive regulation of canonical NF-kappaB signal transduction / intracellular membrane-bounded organelle / identical protein binding ...COPII vesicle coating / COPII-mediated vesicle transport / endoplasmic reticulum exit site / endoplasmic reticulum to Golgi vesicle-mediated transport / bioluminescence / generation of precursor metabolites and energy / Signaling by ALK fusions and activated point mutants / positive regulation of canonical NF-kappaB signal transduction / intracellular membrane-bounded organelle / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||||||||||||||
 Authors Authors | Rosenberg GM / Sawaya MR / Boyer DR / Ge P / Abskharon R / Eisenberg DS | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2023 Journal: PNAS Nexus / Year: 2023Title: Fibril structures of TFG protein mutants validate the identification of TFG as a disease-related amyloid protein by the IMPAcT method. Authors: Gregory M Rosenberg / Romany Abskharon / David R Boyer / Peng Ge / Michael R Sawaya / David S Eisenberg /  Abstract: We previously presented a bioinformatic method for identifying diseases that arise from a mutation in a protein's low-complexity domain that drives the protein into pathogenic amyloid fibrils. One ...We previously presented a bioinformatic method for identifying diseases that arise from a mutation in a protein's low-complexity domain that drives the protein into pathogenic amyloid fibrils. One protein so identified was the tropomyosin-receptor kinase-fused gene protein (TRK-fused gene protein or TFG). Mutations in TFG are associated with degenerative neurological conditions. Here, we present experimental evidence that confirms our prediction that these conditions are amyloid-related. We find that the low-complexity domain of TFG containing the disease-related mutations G269V or P285L forms amyloid fibrils, and we determine their structures using cryo-electron microscopy (cryo-EM). These structures are unmistakably amyloid in nature and confirm the propensity of the mutant TFG low-complexity domain to form amyloid fibrils. Also, despite resulting from a pathogenic mutation, the fibril structures bear some similarities to other amyloid structures that are thought to be nonpathogenic and even functional, but there are other factors that support these structures' relevance to disease, including an increased propensity to form amyloid compared with the wild-type sequence, structure-stabilizing influence from the mutant residues themselves, and double-protofilament amyloid cores. Our findings elucidate two potentially disease-relevant structures of a previously unknown amyloid and also show how the structural features of pathogenic amyloid fibrils may not conform to the features commonly associated with pathogenicity. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41198.map.gz emd_41198.map.gz | 198.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41198-v30.xml emd-41198-v30.xml emd-41198.xml emd-41198.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41198.png emd_41198.png | 149 KB | ||
| Filedesc metadata |  emd-41198.cif.gz emd-41198.cif.gz | 6 KB | ||
| Others |  emd_41198_half_map_1.map.gz emd_41198_half_map_1.map.gz emd_41198_half_map_2.map.gz emd_41198_half_map_2.map.gz | 171.1 MB 171.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41198 http://ftp.pdbj.org/pub/emdb/structures/EMD-41198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41198 | HTTPS FTP |
-Validation report
| Summary document |  emd_41198_validation.pdf.gz emd_41198_validation.pdf.gz | 913.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41198_full_validation.pdf.gz emd_41198_full_validation.pdf.gz | 913.4 KB | Display | |
| Data in XML |  emd_41198_validation.xml.gz emd_41198_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_41198_validation.cif.gz emd_41198_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41198 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41198 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41198 | HTTPS FTP |
-Related structure data
| Related structure data |  8terMC  8teqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41198.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41198.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.539 Å | ||||||||||||||||||||||||||||||||||||
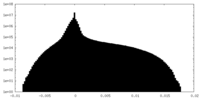
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41198_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
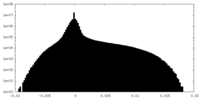
| Density Histograms |
-Half map: #2
| File | emd_41198_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
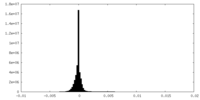
| Density Histograms |
- Sample components
Sample components
-Entire : amyloid fibril of protein TFG P285L
| Entire | Name: amyloid fibril of protein TFG P285L |
|---|---|
| Components |
|
-Supramolecule #1: amyloid fibril of protein TFG P285L
| Supramolecule | Name: amyloid fibril of protein TFG P285L / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: TRK-fused gene protein Low Complexity Domain P285L mutant
| Macromolecule | Name: TRK-fused gene protein Low Complexity Domain P285L mutant type: protein_or_peptide / ID: 1 / Number of copies: 20 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.464754 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMVSKG EEDNMAIIKE FMRFKVHMEG SVNGHEFEIE GEGEGRPYEG TQTAKLKVTK GGPLPFAWD ILSPQFMYGS KAYVKHPADI PDYLKLSFPE GFKWERVMNF EDGGVVTVTQ DSSLQDGEFI YKVKLRGTNF P SDGPVMQK ...String: MSYYHHHHHH DYDIPTTENL YFQGAMVSKG EEDNMAIIKE FMRFKVHMEG SVNGHEFEIE GEGEGRPYEG TQTAKLKVTK GGPLPFAWD ILSPQFMYGS KAYVKHPADI PDYLKLSFPE GFKWERVMNF EDGGVVTVTQ DSSLQDGEFI YKVKLRGTNF P SDGPVMQK KTMGWEASSE RMYPEDGALK GEIKQRLKLK DGGHYDAEVK TTYKAKKPVQ LPGAYNVNIK LDITSHNEDY TI VEQYERA EGRHSTGGMD ELYKAGTMDP GQIEGQMYQQ YQQQAGYGAQ QPQAPPQQPQ QYGIQYSASY SQQTGPQQLQ QFQ GYGQQP TSQAPAPAFS GQPQQLPAQP PQQYQASNYP AQ UniProtKB: Uncharacterized protein, Protein TFG |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 240 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: Quantifoil R1.2/1.3 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV / Details: 8 seconds, blot force 0. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 7920 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 Applied symmetry - Helical parameters - Δz: 2.39 Å Applied symmetry - Helical parameters - Δ&Phi: 179.02 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 2.59 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 193640 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 14.16 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8ter: |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)