[English] 日本語
 Yorodumi
Yorodumi- EMDB-41188: Cryo-EM structure of Arabidopsis thaliana Bor1 mutant (R637E/E641... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Arabidopsis thaliana Bor1 mutant (R637E/E641R/R643E) in the occluded conformation in lauryl maltose neopentyl glycol (LMNG) (protomer-focused refinement) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Borate transporter / Bor1 / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationborate transmembrane transport / response to boron-containing substance / cellular response to boron-containing substance levels / active borate transmembrane transporter activity / monoatomic ion homeostasis / solute:inorganic anion antiporter activity / monoatomic anion transport / vacuolar membrane / transmembrane transport / endosome membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
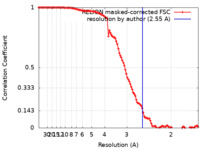
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Jiang Y / Jiang J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of borate transporter Bor1 reveals a novel auto-inhibition mechanism for the SLC4 family Authors: Jiang Y / Jiang J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41188.map.gz emd_41188.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41188-v30.xml emd-41188-v30.xml emd-41188.xml emd-41188.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41188_fsc.xml emd_41188_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41188.png emd_41188.png | 154.2 KB | ||
| Filedesc metadata |  emd-41188.cif.gz emd-41188.cif.gz | 5.6 KB | ||
| Others |  emd_41188_half_map_1.map.gz emd_41188_half_map_1.map.gz emd_41188_half_map_2.map.gz emd_41188_half_map_2.map.gz | 98.3 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41188 http://ftp.pdbj.org/pub/emdb/structures/EMD-41188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41188 | HTTPS FTP |
-Validation report
| Summary document |  emd_41188_validation.pdf.gz emd_41188_validation.pdf.gz | 906 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41188_full_validation.pdf.gz emd_41188_full_validation.pdf.gz | 905.5 KB | Display | |
| Data in XML |  emd_41188_validation.xml.gz emd_41188_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_41188_validation.cif.gz emd_41188_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41188 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41188 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41188 | HTTPS FTP |
-Related structure data
| Related structure data |  8tejMC  8tegC  8tehC  8teiC  8telC  8temC  8tenC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41188.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41188.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
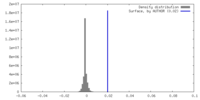
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_41188_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
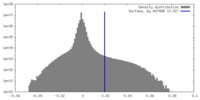
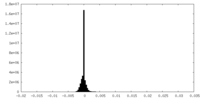
| Density Histograms |
-Half map: #2
| File | emd_41188_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
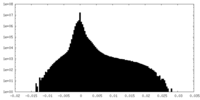
| Density Histograms |
- Sample components
Sample components
-Entire : Arabidopsis thaliana Boron transporter 1
| Entire | Name: Arabidopsis thaliana Boron transporter 1 |
|---|---|
| Components |
|
-Supramolecule #1: Arabidopsis thaliana Boron transporter 1
| Supramolecule | Name: Arabidopsis thaliana Boron transporter 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Boron transporter 1
| Macromolecule | Name: Boron transporter 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 80.108898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEETFVPFEG IKNDLKGRLM CYKQDWTGGF KAGFRILAPT TYIFFASAIP VISFGEQLER STDGVLTAVQ TLASTAICGM IHSIIGGQP LLILGVAEPT VIMYTFMFNF AKARPELGRD LFLAWSGWVC VWTALMLFVL AICGACSIIN RFTRVAGELF G LLIAMLFM ...String: MEETFVPFEG IKNDLKGRLM CYKQDWTGGF KAGFRILAPT TYIFFASAIP VISFGEQLER STDGVLTAVQ TLASTAICGM IHSIIGGQP LLILGVAEPT VIMYTFMFNF AKARPELGRD LFLAWSGWVC VWTALMLFVL AICGACSIIN RFTRVAGELF G LLIAMLFM QQAIKGLVDE FRIPERENQK LKEFLPSWRF ANGMFALVLS FGLLLTGLRS RKARSWRYGT GWLRSLIADY GV PLMVLVW TGVSYIPAGD VPKGIPRRLF SPNPWSPGAY GNWTVVKEML DVPIVYIIGA FIPASMIAVL YYFDHSVASQ LAQ QKEFNL RKPSSYHYDL LLLGFLTLMC GLLGVPPSNG VIPQSPMHTK SLATLKYQLL RNRLVATARR SIKTNASLGQ LYDN MQEAY HHMQ(TPO)PLVYQ QPQGLKELKE STIQATTFTG NLNAPVDETL FDIEKEIDDL LPVEVKEQRV SNLLQSTMVG G CVAAMPIL KMIPTSVLWG YFAFMAIESL PGNQFWERIL LLFTAPSRRF KVLEDYHATF VETVPFKTIA MFTLFQTTYL LI CFGLTWI PIAGVMFPLM IMFLIPVRQY LLPRFFKGAH LQDLDAAEYE EAPALPFNLA AETEIGSTTS YPGDLEILDE VMT ESRGRF EHTSSPKVTS SSSTPVNNRS LSQVFSPRVS GIRLGQMSPR VVGNSPKPAS CGRSPLNQSS SNHHHHHHHH HH UniProtKB: Boron transporter 1 |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 22 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)