+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pom34-Pom152 membrane attachment site yeast NPC | |||||||||||||||
 Map data Map data | c8 ring reconstructed from multibody volume | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nuclear pore complex / nucleocytoplasmic transport / nucleoporin / membrane protein / translocase / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear pore transmembrane ring / spindle pole body duplication / nuclear pore organization / structural constituent of nuclear pore / nucleocytoplasmic transport / nuclear envelope lumen / nuclear outer membrane / mRNA transport / nuclear pore / protein-membrane adaptor activity ...nuclear pore transmembrane ring / spindle pole body duplication / nuclear pore organization / structural constituent of nuclear pore / nucleocytoplasmic transport / nuclear envelope lumen / nuclear outer membrane / mRNA transport / nuclear pore / protein-membrane adaptor activity / nuclear periphery / cell periphery / protein import into nucleus / nuclear envelope / nuclear membrane / mitochondrion Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||||||||
 Authors Authors | Akey CW / Echeverria I / Ouch C / Fernandez-Martinez J / Rout MP | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Implications of a multiscale structure of the yeast nuclear pore complex. Authors: Christopher W Akey / Ignacia Echeverria / Christna Ouch / Ilona Nudelman / Yi Shi / Junjie Wang / Brian T Chait / Andrej Sali / Javier Fernandez-Martinez / Michael P Rout /   Abstract: Nuclear pore complexes (NPCs) direct the nucleocytoplasmic transport of macromolecules. Here, we provide a composite multiscale structure of the yeast NPC, based on improved 3D density maps from ...Nuclear pore complexes (NPCs) direct the nucleocytoplasmic transport of macromolecules. Here, we provide a composite multiscale structure of the yeast NPC, based on improved 3D density maps from cryogenic electron microscopy and AlphaFold2 models. Key features of the inner and outer rings were integrated into a comprehensive model. We resolved flexible connectors that tie together the core scaffold, along with equatorial transmembrane complexes and a lumenal ring that anchor this channel within the pore membrane. The organization of the nuclear double outer ring reveals an architecture that may be shared with ancestral NPCs. Additional connections between the core scaffold and the central transporter suggest that under certain conditions, a degree of local organization is present at the periphery of the transport machinery. These connectors may couple conformational changes in the scaffold to the central transporter to modulate transport. Collectively, this analysis provides insights into assembly, transport, and NPC evolution. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41117.map.gz emd_41117.map.gz | 2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41117-v30.xml emd-41117-v30.xml emd-41117.xml emd-41117.xml | 36.5 KB 36.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41117_fsc.xml emd_41117_fsc.xml | 21.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_41117.png emd_41117.png | 63.6 KB | ||
| Filedesc metadata |  emd-41117.cif.gz emd-41117.cif.gz | 8.5 KB | ||
| Others |  emd_41117_additional_1.map.gz emd_41117_additional_1.map.gz emd_41117_additional_2.map.gz emd_41117_additional_2.map.gz emd_41117_additional_3.map.gz emd_41117_additional_3.map.gz emd_41117_additional_4.map.gz emd_41117_additional_4.map.gz emd_41117_additional_5.map.gz emd_41117_additional_5.map.gz emd_41117_half_map_1.map.gz emd_41117_half_map_1.map.gz emd_41117_half_map_2.map.gz emd_41117_half_map_2.map.gz | 2 MB 378.7 MB 49.9 MB 49.9 MB 4.3 MB 5.3 MB 5.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41117 http://ftp.pdbj.org/pub/emdb/structures/EMD-41117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41117 | HTTPS FTP |
-Validation report
| Summary document |  emd_41117_validation.pdf.gz emd_41117_validation.pdf.gz | 401.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41117_full_validation.pdf.gz emd_41117_full_validation.pdf.gz | 400.7 KB | Display | |
| Data in XML |  emd_41117_validation.xml.gz emd_41117_validation.xml.gz | 24.5 KB | Display | |
| Data in CIF |  emd_41117_validation.cif.gz emd_41117_validation.cif.gz | 33.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41117 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41117 | HTTPS FTP |
-Related structure data
| Related structure data |  8t9lMC  8tieC  8tj5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41117.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41117.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | c8 ring reconstructed from multibody volume | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.66 Å | ||||||||||||||||||||||||||||||||||||
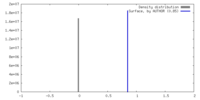
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: extracted TM density for Ndc1
| File | emd_41117_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | extracted TM density for Ndc1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
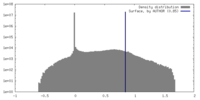
| Density Histograms |
-Additional map: extracted region of micelle ring at lower resolution for the Poms.
| File | emd_41117_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | extracted region of micelle ring at lower resolution for the Poms. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: half map of multibody volume
| File | emd_41117_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of multibody volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
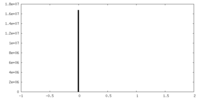
| Density Histograms |
-Additional map: half map of multibody volume
| File | emd_41117_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of multibody volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: c2 map of multibody volume
| File | emd_41117_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | c2 map of multibody volume | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map c8 ring from multibody
| File | emd_41117_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map c8 ring from multibody | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map c8 ring from multibody
| File | emd_41117_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map c8 ring from multibody | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nuclear Pore Complex
| Entire | Name: Nuclear Pore Complex |
|---|---|
| Components |
|
-Supramolecule #1: Nuclear Pore Complex
| Supramolecule | Name: Nuclear Pore Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Protein A tagged Mlp1 pullout of NPC |
|---|---|
| Source (natural) | Organism:  Strain: MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 MLP1-PPX-ProteinA::HIS5 Organelle: nucleus / Location in cell: nuclear envelope |
-Supramolecule #2: Pom34-Pom152 Inner ring anchor complex
| Supramolecule | Name: Pom34-Pom152 Inner ring anchor complex / type: complex / ID: 2 / Parent: 1 Details: native complex resolved by multibody image processing |
|---|---|
| Source (natural) | Organism:  Strain: MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 MLP1-PPX-ProteinA::HIS5 Organelle: nucleus / Location in cell: nuclear envelope |
-Macromolecule #1: Nucleoporin POM152
| Macromolecule | Name: Nucleoporin POM152 / type: protein_or_peptide / ID: 1 Details: Pore outer membrane protein and anchor for the lumenal ring Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 151.833891 KDa |
| Sequence | String: MEHRYNVFND TPRGNHWMGS SVSGSPRPSY SSRPNVNTTR RFQYSDDEPA EKIRPLRSRS FKSTESNISD EKSRISERDS KDRYINGDK KVDIYSLPLI STDVLEISKQ RTFAVILFLI IQCYKIYDLV ILKSGLPLSG LLFKNYRFNF ISKYFIIDSF F LYVLPSFN ...String: MEHRYNVFND TPRGNHWMGS SVSGSPRPSY SSRPNVNTTR RFQYSDDEPA EKIRPLRSRS FKSTESNISD EKSRISERDS KDRYINGDK KVDIYSLPLI STDVLEISKQ RTFAVILFLI IQCYKIYDLV ILKSGLPLSG LLFKNYRFNF ISKYFIIDSF F LYVLPSFN IPRLTFKPWV VYLQILAMLL LNIFISSDHE FVLISLIMTT WRKLYTKELS VTGSAINHHR IFDSSAHFKG AL TIKILPE NTAMFNPLHE SYCLPMDTNL FKINSIDVPI RINSTEEIEY IELEYRDLYT NSVELRSLSK KDFKIIDNPK SFL KKDQSV LKSHSNDFEE GSTIRYLAVT LQDIGFYQIK KIVDSKKLNL KIHQSHLVVP YCPIASITGT GSNDRCIGDS DNVS FEIQG VPPMKLAYSK IVNGQTFSYV DSSLQPEYFE SPLQSSKSKQ SFTQGELNDL KWGRNQPVNI NLDSSITQDG KFAYK IDKI TDGLGNVVDF TSLPEELKKR YDLSYNFNVH EVPRAALEER FDPKSPTKRS IAIVFEEIKN WISDIPYVIS LSYTDA QDK SKKIMNVTTD SLTKVLQADL PGSYNLEYIE SKFCPGEIVG KSNVLVTMPV APTMEVKSFP ILDQCVGQVG LNFELSF TG APPYYYNTKI YKLENGERKL YDAKRYTSEG TRNRFSYSPP KEGNYEIVFD TVSNKLFTEP IKLEPVKEYT FKTSMRVK P SASLKLHHDL KLCLGDHSSV PVALKGQGPF TLTYDIIETF SSKRKTFEIK EIKTNEYVIK TPVFTTGGDY ILSLVSIKD STGCVVGLSQ PDAKIQVRRD IPSAAFNFFE PIKEAKIKHG SVTEIPLKLS GEGPFTVKFK HMDYDGNIVK EFENKFQNSY KPALKVSKE GLYQLVDIRD SSCQGNVIYR NSLYKVSFLE KPKFAIQDNH HITKVTENLF SKEEVCQGME GTVDLALFGS P PFILEYDL MAPNGHISTK KIQVATKYAS LKLPNQIPGE YITTIKAIFD GNYGESDIHF REHQSELIIK QTVHPIPDVA FA DGGKTLR ACAANVDQIS FLEPINLKFL QGESPFSITF SVYHESTSRT DQYTIDNIDS ENFSFEKLYE GMKLGNHAIT IDS VVDANG CVNSLISGPR NQILVSITDA PKIHILDPST EYCVGDYVAY QLNGVAPFMI KYEFNGIPLK SKERSSQFVR LASE PGIIS ITSLQDSSSQ CIVDFTNPKL KSEFDDLSLN IHPIPSVTVS QGNYVTEDIR EGDQAEVIFS FEGTPPFSLT YVRTE ETDG KHGKRRSQVV ETHKVTDIYS HEYKVITSLQ GTYEAIEITD AYCFAKNDLF FNN UniProtKB: Nucleoporin POM152 |
-Macromolecule #2: Nucleoporin POM34
| Macromolecule | Name: Nucleoporin POM34 / type: protein_or_peptide / ID: 2 / Details: Pore membrane protein and inner ring anchor / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.279301 KDa |
| Sequence | String: MKIQAGQLGL DDNDVPGPLP DTDSKPSSQS QNDTPMFKLG NFESPVLKEL SRRTVNKEME TQRIMTNVIA FAFWNLLVKF IKFFWNNTH VGRQFCNRLS RIHLYMLTFH TLKKANIIYH TTFSWLNAEL LDYLFHLLIS LNILFSLWKL LSTVKVSDLN L TDRQKKLL ...String: MKIQAGQLGL DDNDVPGPLP DTDSKPSSQS QNDTPMFKLG NFESPVLKEL SRRTVNKEME TQRIMTNVIA FAFWNLLVKF IKFFWNNTH VGRQFCNRLS RIHLYMLTFH TLKKANIIYH TTFSWLNAEL LDYLFHLLIS LNILFSLWKL LSTVKVSDLN L TDRQKKLL GVDMQSSVDT GLQPQHPHYV STSKISQMAQ NKTHIPQTNL KNHPAYLFKG LETPLKARQR EMAEEQTKLQ SQ SLHTKNV FGTLQRHSGI SSTLVSANND NNSPHTPVTR KGYIPSSKYA YMMNSQSPRG KI UniProtKB: Nucleoporin POM34 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20mM HEPES,50mM Potassium acetate,20mM NaCl,2mM MgCl2,1mM DTT |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK III |
| Details | One step affinity purified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Details | Preliminary grid screening done manually with individual images of low magnification montages of candidate meshes. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-40 / Number grids imaged: 1 / Number real images: 4015 / Average electron dose: 40.0 e/Å2 / Details: 3218 images retained after triage |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated magnification: 37651 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: none |
|---|---|
| Details | Rigid body docking and manual building with Chimera and coot |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: cross correlation |
| Output model |  PDB-8t9l: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)