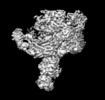
登録情報 データベース : EMDB / ID : EMD-4099タイトル Yeast activated spliceosomal B complex 複合体 : yeast activated spliceosome (BACT)機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Saccharomyces cerevisiae (パン酵母)手法 / / 解像度 : 5.8 Å Stark H / Kastner B / Luehrmann R ジャーナル : Science / 年 : 2016タイトル : Molecular architecture of the Saccharomyces cerevisiae activated spliceosome.著者 : Reinhard Rauhut / Patrizia Fabrizio / Olexandr Dybkov / Klaus Hartmuth / Vladimir Pena / Ashwin Chari / Vinay Kumar / Chung-Tien Lee / Henning Urlaub / Berthold Kastner / Holger Stark / Reinhard Lührmann / 要旨 : The activated spliceosome (B) is in a catalytically inactive state and is remodeled into a catalytically active machine by the RNA helicase Prp2, but the mechanism is unclear. Here, we describe a 3D ... The activated spliceosome (B) is in a catalytically inactive state and is remodeled into a catalytically active machine by the RNA helicase Prp2, but the mechanism is unclear. Here, we describe a 3D electron cryomicroscopy structure of the Saccharomyces cerevisiae B complex at 5.8-angstrom resolution. Our model reveals that in B, the catalytic U2/U6 RNA-Prp8 ribonucleoprotein core is already established, and the 5' splice site (ss) is oriented for step 1 catalysis but occluded by protein. The first-step nucleophile-the branchsite adenosine-is sequestered within the Hsh155 HEAT domain and is held 50 angstroms away from the 5'ss. Our structure suggests that Prp2 adenosine triphosphatase-mediated remodeling leads to conformational changes in Hsh155's HEAT domain that liberate the first-step reactants for catalysis. 履歴 登録 2016年8月17日 - ヘッダ(付随情報) 公開 2016年10月12日 - マップ公開 2016年10月12日 - 更新 2017年7月12日 - 現状 2017年7月12日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 機能・相同性情報
機能・相同性情報
 データ登録者
データ登録者 引用
引用 ジャーナル: Science / 年: 2016
ジャーナル: Science / 年: 2016
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_4099.map.gz
emd_4099.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-4099-v30.xml
emd-4099-v30.xml emd-4099.xml
emd-4099.xml EMDBヘッダ
EMDBヘッダ emd_4099.png
emd_4099.png http://ftp.pdbj.org/pub/emdb/structures/EMD-4099
http://ftp.pdbj.org/pub/emdb/structures/EMD-4099 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4099
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4099 emd_4099_validation.pdf.gz
emd_4099_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_4099_full_validation.pdf.gz
emd_4099_full_validation.pdf.gz emd_4099_validation.xml.gz
emd_4099_validation.xml.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4099
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4099 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4099
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4099 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_4099.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_4099.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素
 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析 ムービー
ムービー コントローラー
コントローラー





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)