[English] 日本語
 Yorodumi
Yorodumi- EMDB-40960: Open-state cryo-EM structure of full-length human TRPV4 in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
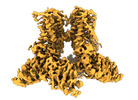
| Title | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | transient receptor potential V family member 4 / TRP / channel / TRPV4 / TRP channels / membrane protein / agonist / 4a-PDD / open state / 4alpha-Phorbol 12 / 13-didecanoate | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationstretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response ...stretch-activated, monoatomic cation-selective, calcium channel activity / blood vessel endothelial cell delamination / regulation of response to osmotic stress / osmosensor activity / vasopressin secretion / calcium ion import into cytosol / positive regulation of microtubule depolymerization / positive regulation of striated muscle contraction / positive regulation of macrophage inflammatory protein 1 alpha production / hyperosmotic salinity response / positive regulation of chemokine (C-X-C motif) ligand 1 production / positive regulation of chemokine (C-C motif) ligand 5 production / cartilage development involved in endochondral bone morphogenesis / cellular hypotonic salinity response / cellular hypotonic response / cortical microtubule organization / multicellular organismal-level water homeostasis / negative regulation of brown fat cell differentiation / positive regulation of vascular permeability / calcium ion import / osmosensory signaling pathway / positive regulation of monocyte chemotactic protein-1 production / cell-cell junction assembly / cellular response to osmotic stress / cell volume homeostasis / regulation of aerobic respiration / TRP channels / cortical actin cytoskeleton / diet induced thermogenesis / positive regulation of macrophage chemotaxis / microtubule polymerization / calcium ion import across plasma membrane / response to mechanical stimulus / alpha-tubulin binding / beta-tubulin binding / monoatomic cation channel activity / cytoplasmic microtubule / SH2 domain binding / actin filament organization / bioluminescence / protein kinase C binding / generation of precursor metabolites and energy / adherens junction / filopodium / positive regulation of JNK cascade / response to insulin / positive regulation of interleukin-6 production / calcium ion transmembrane transport / calcium channel activity / ruffle membrane / intracellular calcium ion homeostasis / positive regulation of inflammatory response / actin filament binding / calcium ion transport / glucose homeostasis / lamellipodium / negative regulation of neuron projection development / cellular response to heat / actin binding / growth cone / positive regulation of cytosolic calcium ion concentration / actin cytoskeleton organization / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / microtubule binding / response to hypoxia / calmodulin binding / positive regulation of ERK1 and ERK2 cascade / apical plasma membrane / cilium / focal adhesion / lipid binding / protein kinase binding / cell surface / negative regulation of transcription by RNA polymerase II / endoplasmic reticulum / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human betaherpesvirus 5 Human betaherpesvirus 5 | |||||||||||||||||||||
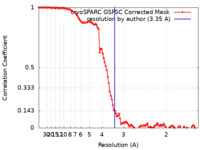
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||||||||||||||
 Authors Authors | Talyzina IA / Nadezhdin KD / Neuberger A / Sobolevsky AI | |||||||||||||||||||||
| Funding support |  United States, United States,  Germany, 6 items Germany, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of human TRPV4 in complex with GTPase RhoA. Authors: Kirill D Nadezhdin / Irina A Talyzina / Aravind Parthasarathy / Arthur Neuberger / David X Zhang / Alexander I Sobolevsky /  Abstract: Transient receptor potential (TRP) channel TRPV4 is a polymodal cellular sensor that responds to moderate heat, cell swelling, shear stress, and small-molecule ligands. It is involved in ...Transient receptor potential (TRP) channel TRPV4 is a polymodal cellular sensor that responds to moderate heat, cell swelling, shear stress, and small-molecule ligands. It is involved in thermogenesis, regulation of vascular tone, bone homeostasis, renal and pulmonary functions. TRPV4 is implicated in neuromuscular and skeletal disorders, pulmonary edema, and cancers, and represents an important drug target. The cytoskeletal remodeling GTPase RhoA has been shown to suppress TRPV4 activity. Here, we present a structure of the human TRPV4-RhoA complex that shows RhoA interaction with the membrane-facing surface of the TRPV4 ankyrin repeat domains. The contact interface reveals residues that are mutated in neuropathies, providing an insight into the disease pathogenesis. We also identify the binding sites of the TRPV4 agonist 4α-PDD and the inhibitor HC-067047 at the base of the S1-S4 bundle, and show that agonist binding leads to pore opening, while channel inhibition involves a π-to-α transition in the pore-forming helix S6. Our structures elucidate the interaction interface between hTRPV4 and RhoA, as well as residues at this interface that are involved in TRPV4 disease-causing mutations. They shed light on TRPV4 activation and inhibition and provide a template for the design of future therapeutics for treatment of TRPV4-related diseases. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40960.map.gz emd_40960.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40960-v30.xml emd-40960-v30.xml emd-40960.xml emd-40960.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40960_fsc.xml emd_40960_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40960.png emd_40960.png | 106.4 KB | ||
| Filedesc metadata |  emd-40960.cif.gz emd-40960.cif.gz | 7 KB | ||
| Others |  emd_40960_half_map_1.map.gz emd_40960_half_map_1.map.gz emd_40960_half_map_2.map.gz emd_40960_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40960 http://ftp.pdbj.org/pub/emdb/structures/EMD-40960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40960 | HTTPS FTP |
-Related structure data
| Related structure data |  8t1dMC  8t1bC  8t1cC  8t1eC  8t1fC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40960.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40960.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7888 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40960_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40960_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : full-length human TRPV4 in complex with agonist 4a-PDD
| Entire | Name: full-length human TRPV4 in complex with agonist 4a-PDD |
|---|---|
| Components |
|
-Supramolecule #1: full-length human TRPV4 in complex with agonist 4a-PDD
| Supramolecule | Name: full-length human TRPV4 in complex with agonist 4a-PDD type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 4/...
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
| Molecular weight | Theoretical: 127.717938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG ...String: MADSSEGPRA GPGEVAELPG DESGTPGGEA FPLSSLANLF EGEDGSLSPS PADASRPAGP GDGRPNLRMK FQGAFRKGVP NPIDLLEST LYESSVVPGP KKAPMDSLFD YGTYRHHSSD NKRWRKKIIE KQPQSPKAPA PQPPPILKVF NRPILFDIVS R GSTADLDG LLPFLLTHKK RLTDEEFREP STGKTCLPKA LLNLSNGRND TIPVLLDIAE RTGNMREFIN SPFRDIYYRG QT ALHIAIE RRCKHYVELL VAQGADVHAQ ARGRFFQPKD EGGYFYFGEL PLSLAACTNQ PHIVNYLTEN PHKKADMRRQ DSR GNTVLH ALVAIADNTR ENTKFVTKMY DLLLLKCARL FPDSNLEAVL NNDGLSPLMM AAKTGKIGIF QHIIRREVTD EDTR HLSRK FKDWAYGPVY SSLYDLSSLD TCGEEASVLE ILVYNSKIEN RHEMLAVEPI NELLRDKWRK FGAVSFYINV VSYLC AMVI FTLTAYYQPL EGTPPYPYRT TVDYLRLAGE VITLFTGVLF FFTNIKDLFM KKCPGVNSLF IDGSFQLLYF IYSVLV IVS AALYLAGIEA YLAVMVFALV LGWMNALYFT RGLKLTGTYS IMIQKILFKD LFRFLLVYLL FMIGYASALV SLLNPCA NM KVCNEDQTNC TVPTYPSCRD SETFSTFLLD LFKLTIGMGD LEMLSSTKYP VVFIILLVTY IILTFVLLLN MLIALMGE T VGQVSKESKH IWKLQWATTI LDIERSFPVF LRKAFRSGEM VTVGKSSDGT PDRRWCFRVD EVNWSHWNQN LGIINEDPG KNETYQYYGF SHTVGRLRRD RWSSVVPRVV ELNKNSNPDE VVVPLDSMGN PRCDGHQQGY PRKWRTDDAP LLVPRGSAAA AVSKGEELF TGVVPILVEL DGDVNGHKFS VSGEGEGDAT YGKLTLKFIC TTGKLPVPWP TLVTTLTYGV QCFSRYPDHM K QHDFFKSA MPEGYVQERT IFFKDDGNYK TRAEVKFEGD TLVNRIELKG IDFKEDGNIL GHKLEYNYNS HNVYIMADKQ KN GIKVNFK IRHNIEDGSV QLADHYQQNT PIGDGPVLLP DNHYLSTQSK LSKDPNEKRD HMVLLEFVTA AGITLGMDEL YKS GLRSWS HPQFEK UniProtKB: Transient receptor potential cation channel subfamily V member 4, Enhanced green fluorescent protein |
-Macromolecule #2: (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-...
| Macromolecule | Name: (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate type: ligand / ID: 2 / Number of copies: 4 / Formula: XS9 |
|---|---|
| Molecular weight | Theoretical: 672.931 Da |
| Chemical component information | 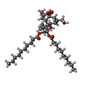 ChemComp-XS9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | human TRPV4 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 15624 / Average exposure time: 2.4 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8t1d: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)