+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4096 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Yvh1 pre-60S particle | |||||||||
 Map data Map data | Yvh1 pre-60S particle | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||
 Authors Authors | Sarkar A / Pech M / Thoms M / Beckmann R / Hurt E | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2016 Journal: Nat Struct Mol Biol / Year: 2016Title: Ribosome-stalk biogenesis is coupled with recruitment of nuclear-export factor to the nascent 60S subunit. Authors: Anshuk Sarkar / Markus Pech / Matthias Thoms / Roland Beckmann / Ed Hurt /  Abstract: Nuclear export of preribosomal subunits is a key step during eukaryotic ribosome formation. To efficiently pass through the FG-repeat meshwork of the nuclear pore complex, the large pre-60S subunit ...Nuclear export of preribosomal subunits is a key step during eukaryotic ribosome formation. To efficiently pass through the FG-repeat meshwork of the nuclear pore complex, the large pre-60S subunit requires several export factors. Here we describe the mechanism of recruitment of the Saccharomyces cerevisiae RNA-export receptor Mex67-Mtr2 to the pre-60S subunit at the proper time. Mex67-Mtr2 binds at the premature ribosomal-stalk region, which later during translation serves as a binding platform for translational GTPases on the mature ribosome. The assembly factor Mrt4, a structural homolog of cytoplasmic-stalk protein P0, masks this site, thus preventing untimely recruitment of Mex67-Mtr2 to nuclear pre-60S particles. Subsequently, Yvh1 triggers Mrt4 release in the nucleus, thereby creating a narrow time window for Mex67-Mtr2 association at this site and facilitating nuclear export of the large subunit. Thus, a spatiotemporal mark on the ribosomal stalk controls the recruitment of an RNA-export receptor to the nascent 60S subunit. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4096.map.gz emd_4096.map.gz | 264.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4096-v30.xml emd-4096-v30.xml emd-4096.xml emd-4096.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4096.png emd_4096.png | 54 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4096 http://ftp.pdbj.org/pub/emdb/structures/EMD-4096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4096 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4096.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4096.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yvh1 pre-60S particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.062 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
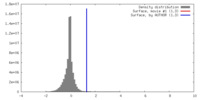
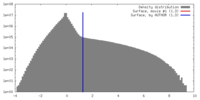
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yvh1 pre-60S particle
| Entire | Name: Yvh1 pre-60S particle |
|---|---|
| Components |
|
-Supramolecule #1: Yvh1 pre-60S particle
| Supramolecule | Name: Yvh1 pre-60S particle / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK IV / Details: Method Blot for 3 seconds before plunging. | ||||||||||||
| Details | The freshly prepared pre-60S sample was applied to 2 nm pre-coated Quantifoil holey carbon supparted grids an vitrified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 3226 / Average electron dose: 4.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.66 µm / Nominal defocus min: 1.06 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)