[English] 日本語
 Yorodumi
Yorodumi- EMDB-40907: Cryo-EM Structure of NINJ2 Filament at 3.07 Angstrom Resolution -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of NINJ2 Filament at 3.07 Angstrom Resolution | |||||||||
 Map data Map data | Cryo-EM Map of NINJ2 Filament | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NINJ2 Filament / NINJ1 Paralog Protein / Cholesterol Binding Protein / Lipid Binding Protein / MEMBRANE PROTEIN | |||||||||
| Function / homology | Ninjurin / Ninjurin / neuron cell-cell adhesion / tissue regeneration / cholesterol binding / nervous system development / cell adhesion / plasma membrane / Ninjurin-2 Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
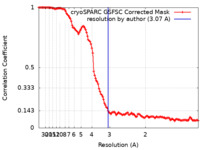
| Method | single particle reconstruction / cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Sahoo B / Dai X | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2025 Journal: Cell / Year: 2025Title: How NINJ1 mediates plasma membrane rupture and why NINJ2 cannot. Authors: Bibekananda Sahoo / Zongjun Mou / Wei Liu / George Dubyak / Xinghong Dai /  Abstract: Ninjurin-1 (NINJ1) is an active executioner of plasma membrane rupture (PMR), a process previously thought to be a passive osmotic lysis event in lytic cell death. Ninjurin-2 (NINJ2) is a close ...Ninjurin-1 (NINJ1) is an active executioner of plasma membrane rupture (PMR), a process previously thought to be a passive osmotic lysis event in lytic cell death. Ninjurin-2 (NINJ2) is a close paralog of NINJ1 but cannot mediate PMR. Using cryogenic electron microscopy (cryo-EM), we show that NINJ1 and NINJ2 both assemble into linear filaments that are hydrophobic on one side but hydrophilic on the other. This structural feature and other evidence point to a PMR mechanism by which NINJ1 filaments wrap around and solubilize membrane fragments and, less frequently, form pores in the plasma membrane. In contrast to the straight NINJ1 filament, the NINJ2 filament is curved toward the intracellular space, preventing its circularization or even assembly on a relatively flat membrane to mediate PMR. Mutagenesis studies further demonstrate that the NINJ2 filament curvature is induced by strong association with lipids, particularly a cholesterol molecule, at the cytoplasmic leaflet of the lipid bilayer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40907.map.gz emd_40907.map.gz | 167.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40907-v30.xml emd-40907-v30.xml emd-40907.xml emd-40907.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40907_fsc.xml emd_40907_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_40907.png emd_40907.png | 83 KB | ||
| Filedesc metadata |  emd-40907.cif.gz emd-40907.cif.gz | 5.7 KB | ||
| Others |  emd_40907_half_map_1.map.gz emd_40907_half_map_1.map.gz emd_40907_half_map_2.map.gz emd_40907_half_map_2.map.gz | 165.3 MB 165.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40907 http://ftp.pdbj.org/pub/emdb/structures/EMD-40907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40907 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40907 | HTTPS FTP |
-Validation report
| Summary document |  emd_40907_validation.pdf.gz emd_40907_validation.pdf.gz | 912.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40907_full_validation.pdf.gz emd_40907_full_validation.pdf.gz | 911.9 KB | Display | |
| Data in XML |  emd_40907_validation.xml.gz emd_40907_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_40907_validation.cif.gz emd_40907_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40907 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40907 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40907 | HTTPS FTP |
-Related structure data
| Related structure data |  8szbMC  8szaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40907.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40907.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Map of NINJ2 Filament | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM Half Map A of NINJ2 Filament
| File | emd_40907_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Half Map A of NINJ2 Filament | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM Half Map B of NINJ2 Filament
| File | emd_40907_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM Half Map B of NINJ2 Filament | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ninjurin-2 in complex with Cholesterol
| Entire | Name: Ninjurin-2 in complex with Cholesterol |
|---|---|
| Components |
|
-Supramolecule #1: Ninjurin-2 in complex with Cholesterol
| Supramolecule | Name: Ninjurin-2 in complex with Cholesterol / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Ninjurin-2
| Macromolecule | Name: Ninjurin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.615115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKGSGESAR ENIDLQPGSS DPRSQPINLN HYATKKSVAE SMLDVALFMS NAMRLKAVLE QGPSSHYYT TLVTLISLSL LLQVVIGVLL VVIARLNLNE VEKQWRLNQL NNAATILVFF TVVINVFITA FGAHKTGFLA A RASRNPL UniProtKB: Ninjurin-2 |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 6 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)