[English] 日本語
 Yorodumi
Yorodumi- EMDB-4085: The Dimeric Architecture of Checkpoint Kinases Mec1/ATR and Tel1/... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4085 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
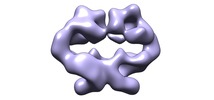
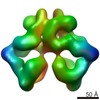
| Title | The Dimeric Architecture of Checkpoint Kinases Mec1/ATR and Tel1/ATM Reveal a Common Structural Organization. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 22.5 Å | |||||||||
 Authors Authors | Sawicka M / Wanrooij PH / Darbari VC / Tannous E / Hailemariam S / Bose D / Makarova AV / Burgers PM / Zhang X | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2016 Journal: J Biol Chem / Year: 2016Title: The Dimeric Architecture of Checkpoint Kinases Mec1ATR and Tel1ATM Reveal a Common Structural Organization. Authors: Marta Sawicka / Paulina H Wanrooij / Vidya C Darbari / Elias Tannous / Sarem Hailemariam / Daniel Bose / Alena V Makarova / Peter M Burgers / Xiaodong Zhang /   Abstract: The phosphatidylinositol 3-kinase-related protein kinases are key regulators controlling a wide range of cellular events. The yeast Tel1 and Mec1·Ddc2 complex (ATM and ATR-ATRIP in humans) play ...The phosphatidylinositol 3-kinase-related protein kinases are key regulators controlling a wide range of cellular events. The yeast Tel1 and Mec1·Ddc2 complex (ATM and ATR-ATRIP in humans) play pivotal roles in DNA replication, DNA damage signaling, and repair. Here, we present the first structural insight for dimers of Mec1·Ddc2 and Tel1 using single-particle electron microscopy. Both kinases reveal a head to head dimer with one major dimeric interface through the N-terminal HEAT (named after Huntingtin, elongation factor 3, protein phosphatase 2A, and yeast kinase TOR1) repeat. Their dimeric interface is significantly distinct from the interface of mTOR complex 1 dimer, which oligomerizes through two spatially separate interfaces. We also observe different structural organizations of kinase domains of Mec1 and Tel1. The kinase domains in the Mec1·Ddc2 dimer are located in close proximity to each other. However, in the Tel1 dimer they are fully separated, providing potential access of substrates to this kinase, even in its dimeric form. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4085.map.gz emd_4085.map.gz | 363.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4085-v30.xml emd-4085-v30.xml emd-4085.xml emd-4085.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4085.png emd_4085.png | 128.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4085 http://ftp.pdbj.org/pub/emdb/structures/EMD-4085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4085 | HTTPS FTP |
-Validation report
| Summary document |  emd_4085_validation.pdf.gz emd_4085_validation.pdf.gz | 189.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4085_full_validation.pdf.gz emd_4085_full_validation.pdf.gz | 188.2 KB | Display | |
| Data in XML |  emd_4085_validation.xml.gz emd_4085_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4085 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4085 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4085 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4085 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4085.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4085.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Checkpoint Kinase Mec1 with its associating protein Ddc2
| Entire | Name: Complex of Checkpoint Kinase Mec1 with its associating protein Ddc2 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Checkpoint Kinase Mec1 with its associating protein Ddc2
| Supramolecule | Name: Complex of Checkpoint Kinase Mec1 with its associating protein Ddc2 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 720 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
| Staining | Type: NEGATIVE / Material: 2% v/v uranyl acetate | ||||||||||||||||||
| Grid | Model: TAAB / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 3) |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 22.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.3) / Number images used: 5633 |
| Initial angle assignment | Type: COMMON LINE / Software - Name: IMAGIC (ver. V) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: IMAGIC (ver. V) |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















