+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4069 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | RadA dodecamer on ssDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
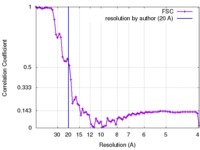
| Method | single particle reconstruction / negative staining / Resolution: 20.0 Å | |||||||||
 Authors Authors | Rapisarda C / Fronzes R | |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: RadA dodecamer on ssDNA Authors: Rapisarda C / Fronzes R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4069.map.gz emd_4069.map.gz | 21.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4069-v30.xml emd-4069-v30.xml emd-4069.xml emd-4069.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4069_fsc.xml emd_4069_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_4069.png emd_4069.png | 43 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4069 http://ftp.pdbj.org/pub/emdb/structures/EMD-4069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4069 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4069.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4069.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.99 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
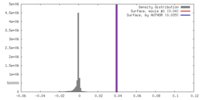
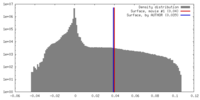
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of RadA with ssDNA
| Entire | Name: Complex of RadA with ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: Complex of RadA with ssDNA
| Supramolecule | Name: Complex of RadA with ssDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: RadA
| Macromolecule | Name: RadA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MASWSHPQFE KSGGGGGLVP RGSKKKATFV CQNCGYNSPK YLGRCPNCGS WSSFVEEVEV AEVKNARVSL TGEKTKPMKL AEVTSINVNR TKTEMEEFNR VLGGGVVPGS LVLIGGDPGI GKSTLLLQVS TQLSQVGTVL YVSGEESAQQ IKLRAERLGD IDSEFYLYAE ...String: MASWSHPQFE KSGGGGGLVP RGSKKKATFV CQNCGYNSPK YLGRCPNCGS WSSFVEEVEV AEVKNARVSL TGEKTKPMKL AEVTSINVNR TKTEMEEFNR VLGGGVVPGS LVLIGGDPGI GKSTLLLQVS TQLSQVGTVL YVSGEESAQQ IKLRAERLGD IDSEFYLYAE TNMQSVRAEV ERIQPDFLII DSIQTIMSPE ISGVQGSVSQ VREVTAELMQ LAKTNNIAIF IVGHVTKEGT LAGPRMLEHM VDTVLYFEGE RHHTFRILRA VKNRFGSTNE IGIFEMQSGG LVEVLNPSQV FLEERLDGAT GSSIVVTMEG TRPILAEVQA LVTPTMFGNA KRTTTGLDFN RASLIMAVLE KRAGLLLQNQ DAYLKSAGGV KLDEPAIDLA VAVAIASSYK DKPTNPQECF VGELGLTGEI RRVNRIEQRI NEAAKLGFTK IYVPKNSLTG ITLPKEIQVI GVTTIQEVLK KVF |
-Macromolecule #2: M13mp18
| Macromolecule | Name: M13mp18 / type: dna / ID: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: AATGCTACTA CTATTAGTAG AATTGATGCC ACCTTTTCAG CTCGCGCCCC AAATGAAAAT ATAGCTAAAC AGGTTATTGA CCATTTGCG AAATGTATCT AATGGTCAAA CTAAATCTAC TCGTTCGCAG AATTGGGAAT CAACTGTTAT ATGGAATGAA A CTTCCAGA ...String: AATGCTACTA CTATTAGTAG AATTGATGCC ACCTTTTCAG CTCGCGCCCC AAATGAAAAT ATAGCTAAAC AGGTTATTGA CCATTTGCG AAATGTATCT AATGGTCAAA CTAAATCTAC TCGTTCGCAG AATTGGGAAT CAACTGTTAT ATGGAATGAA A CTTCCAGA CACCGTACTT TAGTTGCATA TTTAAAACAT GTTGAGCTAC AGCATTATAT TCAGCAATTA AGCTCTAAGC CA TCCGCAA AAATGACCTC TTATCAAAAG GAGCAATTAA AGGTACTCTC TAATCCTGAC CTGTTGGAGT TTGCTTCCGG TCT GGTTCG CTTTGAAGCT CGAATTAAAA CGCGATATTT GAAGTCTTTC GGGCTTCCTC TTAATCTTTT TGATGCAATC CGCT TTGCT TCTGACTATA ATAGTCAGGG TAAAGACCTG ATTTTTGATT TATGGTCATT CTCGTTTTCT GAACTGTTTA AAGCA TTTG AGGGGGATTC AATGAATATT TATGACGATT CCGCAGTATT GGACGCTATC CAGTCTAAAC ATTTTACTAT TACCCC CTC TGGCAAAACT TCTTTTGCAA AAGCCTCTCG CTATTTTGGT TTTTATCGTC GTCTGGTAAA CGAGGGTTAT GATAGTG TT GCTCTTACTA TGCCTCGTAA TTCCTTTTGG CGTTATGTAT CTGCATTAGT TGAATGTGGT ATTCCTAAAT CTCAACTG A TGAATCTTTC TACCTGTAAT AATGTTGTTC CGTTAGTTCG TTTTATTAAC GTAGATTTTT CTTCCCAACG TCCTGACTG GTATAATGAG CCAGTTCTTA AAATCGCATA AGGTAATTCA CAATGATTAA AGTTGAAATT AAACCATCTC AAGCCCAATT TACTACTCG TTCTGGTGTT TCTCGTCAGG GCAAGCCTTA TTCACTGAAT GAGCAGCTTT GTTACGTTGA TTTGGGTAAT G AATATCCG GTTCTTGTCA AGATTACTCT TGATGAAGGT CAGCCAGCCT ATGCGCCTGG TCTGTACACC GTTCATCTGT CC TCTTTCA AAGTTGGTCA GTTCGGTTCC CTTATGATTG ACCGTCTGCG CCTCGTTCCG GCTAAGTAAC ATGGAGCAGG TCG CGGATT TCGACACAAT TTATCAGGCG ATGATACAAA TCTCCGTTGT ACTTTGTTTC GCGCTTGGTA TAATCGCTGG GGGT CAAAG ATGAGTGTTT TAGTGTATTC TTTTGCCTCT TTCGTTTTAG GTTGGTGCCT TCGTAGTGGC ATTACGTATT TTACC CGTT TAATGGAAAC TTCCTCATGA AAAAGTCTTT AGTCCTCAAA GCCTCTGTAG CCGTTGCTAC CCTCGTTCCG ATGCTG TCT TTCGCTGCTG AGGGTGACGA TCCCGCAAAA GCGGCCTTTA ACTCCCTGCA AGCCTCAGCG ACCGAATATA TCGGTTA TG CGTGGGCGAT GGTTGTTGTC ATTGTCGGCG CAACTATCGG TATCAAGCTG TTTAAGAAAT TCACCTCGAA AGCAAGCT G ATAAACCGAT ACAATTAAAG GCTCCTTTTG GAGCCTTTTT TTTGGAGATT TTCAACGTGA AAAAATTATT ATTCGCAAT TCCTTTAGTT GTTCCTTTCT ATTCTCACTC CGCTGAAACT GTTGAAAGTT GTTTAGCAAA ATCCCATACA GAAAATTCAT TTACTAACG TCTGGAAAGA CGACAAAACT TTAGATCGTT ACGCTAACTA TGAGGGCTGT CTGTGGAATG CTACAGGCGT T GTAGTTTG TACTGGTGAC GAAACTCAGT GTTACGGTAC ATGGGTTCCT ATTGGGCTTG CTATCCCTGA AAATGAGGGT GG TGGCTCT GAGGGTGGCG GTTCTGAGGG TGGCGGTTCT GAGGGTGGCG GTACTAAACC TCCTGAGTAC GGTGATACAC CTA TTCCGG GCTATACTTA TATCAACCCT CTCGACGGCA CTTATCCGCC TGGTACTGAG CAAAACCCCG CTAATCCTAA TCCT TCTCT TGAGGAGTCT CAGCCTCTTA ATACTTTCAT GTTTCAGAAT AATAGGTTCC GAAATAGGCA GGGGGCATTA ACTGT TTAT ACGGGCACTG TTACTCAAGG CACTGACCCC GTTAAAACTT ATTACCAGTA CACTCCTGTA TCATCAAAAG CCATGT ATG ACGCTTACTG GAACGGTAAA TTCAGAGACT GCGCTTTCCA TTCTGGCTTT AATGAGGATT TATTTGTTTG TGAATAT CA AGGCCAATCG TCTGACCTGC CTCAACCTCC TGTCAATGCT GGCGGCGGCT CTGGTGGTGG TTCTGGTGGC GGCTCTGA G GGTGGTGGCT CTGAGGGTGG CGGTTCTGAG GGTGGCGGCT CTGAGGGAGG CGGTTCCGGT GGTGGCTCTG GTTCCGGTG ATTTTGATTA TGAAAAGATG GCAAACGCTA ATAAGGGGGC TATGACCGAA AATGCCGATG AAAACGCGCT ACAGTCTGAC GCTAAAGGC AAACTTGATT CTGTCGCTAC TGATTACGGT GCTGCTATCG ATGGTTTCAT TGGTGACGTT TCCGGCCTTG C TAATGGTA ATGGTGCTAC TGGTGATTTT GCTGGCTCTA ATTCCCAAAT GGCTCAAGTC GGTGACGGTG ATAATTCACC TT TAATGAA TAATTTCCGT CAATATTTAC CTTCCCTCCC TCAATCGGTT GAATGTCGCC CTTTTGTCTT TGGCGCTGGT AAA CCATAT GAATTTTCTA TTGATTGTGA CAAAATAAAC TTATTCCGTG GTGTCTTTGC GTTTCTTTTA TATGTTGCCA CCTT TATGT ATGTATTTTC TACGTTTGCT AACATACTGC GTAATAAGGA GTCTTAATCA TGCCAGTTCT TTTGGGTATT CCGTT ATTA TTGCGTTTCC TCGGTTTCCT TCTGGTAACT TTGTTCGGCT ATCTGCTTAC TTTTCTTAAA AAGGGCTTCG GTAAGA TAG CTATTGCTAT TTCATTGTTT CTTGCTCTTA TTATTGGGCT TAACTCAATT CTTGTGGGTT ATCTCTCTGA TATTAGC GC TCAATTACCC TCTGACTTTG TTCAGGGTGT TCAGTTAATT CTCCCGTCTA ATGCGCTTCC CTGTTTTTAT GTTATTCT C TCTGTAAAGG CTGCTATTTT CATTTTTGAC GTTAAACAAA AAATCGTTTC TTATTTGGAT TGGGATAAAT AATATGGCT GTTTATTTTG TAACTGGCAA ATTAGGCTCT GGAAAGACGC TCGTTAGCGT TGGTAAGATT CAGGATAAAA TTGTAGCTGG GTGCAAAAT AGCAACTAAT CTTGATTTAA GGCTTCAAAA CCTCCCGCAA GTCGGGAGGT TCGCTAAAAC GCCTCGCGTT C TTAGAATA CCGGATAAGC CTTCTATATC TGATTTGCTT GCTATTGGGC GCGGTAATGA TTCCTACGAT GAAAATAAAA AC GGCTTGC TTGTTCTCGA TGAGTGCGGT ACTTGGTTTA ATACCCGTTC TTGGAATGAT AAGGAAAGAC AGCCGATTAT TGA TTGGTT TCTACATGCT CGTAAATTAG GATGGGATAT TATTTTTCTT GTTCAGGACT TATCTATTGT TGATAAACAG GCGC GTTCT GCATTAGCTG AACATGTTGT TTATTGTCGT CGTCTGGACA GAATTACTTT ACCTTTTGTC GGTACTTTAT ATTCT CTTA TTACTGGCTC GAAAATGCCT CTGCCTAAAT TACATGTTGG CGTTGTTAAA TATGGCGATT CTCAATTAAG CCCTAC TGT TGAGCGTTGG CTTTATACTG GTAAGAATTT GTATAACGCA TATGATACTA AACAGGCTTT TTCTAGTAAT TATGATT CC GGTGTTTATT CTTATTTAAC GCCTTATTTA TCACACGGTC GGTATTTCAA ACCATTAAAT TTAGGTCAGA AGATGAAA T TAACTAAAAT ATATTTGAAA AAGTTTTCTC GCGTTCTTTG TCTTGCGATT GGATTTGCAT CAGCATTTAC ATATAGTTA TATAACCCAA CCTAAGCCGG AGGTTAAAAA GGTAGTCTCT CAGACCTATG ATTTTGATAA ATTCACTATT GACTCTTCTC AGCGTCTTA ATCTAAGCTA TCGCTATGTT TTCAAGGATT CTAAGGGAAA ATTAATTAAT AGCGACGATT TACAGAAGCA A GGTTATTC ACTCACATAT ATTGATTTAT GTACTGTTTC CATTAAAAAA GGTAATTCAA ATGAAATTGT TAAATGTAAT TA ATTTTGT TTTCTTGATG TTTGTTTCAT CATCTTCTTT TGCTCAGGTA ATTGAAATGA ATAATTCGCC TCTGCGCGAT TTT GTAACT TGGTATTCAA AGCAATCAGG CGAATCCGTT ATTGTTTCTC CCGATGTAAA AGGTACTGTT ACTGTATATT CATC TGACG TTAAACCTGA AAATCTACGC AATTTCTTTA TTTCTGTTTT ACGTGCAAAT AATTTTGATA TGGTAGGTTC TAACC CTTC CATTATTCAG AAGTATAATC CAAACAATCA GGATTATATT GATGAATTGC CATCATCTGA TAATCAGGAA TATGAT GAT AATTCCGCTC CTTCTGGTGG TTTCTTTGTT CCGCAAAATG ATAATGTTAC TCAAACTTTT AAAATTAATA ACGTTCG GG CAAAGGATTT AATACGAGTT GTCGAATTGT TTGTAAAGTC TAATACTTCT AAATCCTCAA ATGTATTATC TATTGACG G CTCTAATCTA TTAGTTGTTA GTGCTCCTAA AGATATTTTA GATAACCTTC CTCAATTCCT TTCAACTGTT GATTTGCCA ACTGACCAGA TATTGATTGA GGGTTTGATA TTTGAGGTTC AGCAAGGTGA TGCTTTAGAT TTTTCATTTG CTGCTGGCTC TCAGCGTGG CACTGTTGCA GGCGGTGTTA ATACTGACCG CCTCACCTCT GTTTTATCTT CTGCTGGTGG TTCGTTCGGT A TTTTTAAT GGCGATGTTT TAGGGCTATC AGTTCGCGCA TTAAAGACTA ATAGCCATTC AAAAATATTG TCTGTGCCAC GT ATTCTTA CGCTTTCAGG TCAGAAGGGT TCTATCTCTG TTGGCCAGAA TGTCCCTTTT ATTACTGGTC GTGTGACTGG TGA ATCTGC CAATGTAAAT AATCCATTTC AGACGATTGA GCGTCAAAAT GTAGGTATTT CCATGAGCGT TTTTCCTGTT GCAA TGGCT GGCGGTAATA TTGTTCTGGA TATTACCAGC AAGGCCGATA GTTTGAGTTC TTCTACTCAG GCAAGTGATG TTATT ACTA ATCAAAGAAG TATTGCTACA ACGGTTAATT TGCGTGATGG ACAGACTCTT TTACTCGGTG GCCTCACTGA TTATAA AAA CACTTCTCAG GATTCTGGCG TACCGTTCCT GTCTAAAATC CCTTTAATCG GCCTCCTGTT TAGCTCCCGC TCTGATT CT AACGAGGAAA GCACGTTATA CGTGCTCGTC AAAGCAACCA TAGTACGCGC CCTGTAGCGG CGCATTAAGC GCGGCGGG T GTGGTGGTTA CGCGCAGCGT GACCGCTACA CTTGCCAGCG CCCTAGCGCC CGCTCCTTTC GCTTTCTTCC CTTCCTTTC TCGCCACGTT CGCCGGCTTT CCCCGTCAAG CTCTAAATCG GGGGCTCCCT TTAGGGTTCC GATTTAGTGC TTTACGGCAC CTCGACCCC AAAAAACTTG ATTTGGGTGA TGGTTCACGT AGTGGGCCAT CGCCCTGATA GACGGTTTTT CGCCCTTTGA C GTTGGAGT CCACGTTCTT TAATAGTGGA CTCTTGTTCC AAACTGGAAC AACACTCAAC CCTATCTCGG GCTATTCTTT TG ATTTATA AGGGATTTTG CCGATTTCGG AACCACCATC AAACAGGATT TTCGCCTGCT GGGGCAAACC AGCGTGGACC GCT TGCTGC AACTCTCTCA GGGCCAGGCG GTGAAGGGCA ATCAGCTGTT GCCCGTCTCA CTGGTGAAAA GAAAAACCAC CCTG GCGCC CAATACGCAA ACCGCCTCTC CCCGCGCGTT GGCCGATTCA TTAATGCAGC TGGCACGACA GGTTTCCCGA CTGGA AAGC GGGCAGTGAG CGCAACGCAA TTAATGTGAG TTAGCTCACT CATTAGGCAC CCCAGGCTTT ACACTTTATG CTTCCG GCT CGTATGTTGT GTGGAATTGT GAGCGGATAA CAATTTCACA CAGGAAACAG CTATGACCAT GATTACGAAT TCGAGCT CG GTACCCGGGG ATCCTCTAGA GTCGACCTGC AGGCATGCAA GCTTGGCACT GGCCGTCGTT TTACAACGTC GTGACTGG G AAAACCCTGG CGTTACCCAA CTTAATCGCC TTGCAGCACA TCCCCCTTTC GCCAGCTGGC GTAATAGCGA AGAGGCCCG CACCGATCGC CCTTCCCAAC AGTTGCGCAG CCTGAATGGC GAATGGCGCT TTGCCTGGTT TCCGGCACCA GAAGCGGTGC CGGAAAGCT GGCTGGAGTG CGATCTTCCT GAGGCCGATA CTGTCGTCGT CCCCTCAAAC TGGCAGATGC ACGGTTACGA T GCGCCCAT CTACACCAAC GTGACCTATC CCATTACGGT CAATCCGCCG TTTGTTCCCA CGGAGAATCC GACGGGTTGT TA CTCGCTC ACATTTAATG TTGATGAAAG CTGGCTACAG GAAGGCCAGA CGCGAATTAT TTTTGATGGC GTTCCTATTG GTT AAAAAA TGAGCTGATT TAACAAAAAT TTAATGCGAA TTTTAACAAA ATATTAACGT TTACAATTTA AATATTTGCT TATA CAATC TTCCTGTTTT TGGGGCTTTT CTGATTATCA ACCGGGGTAC ATATGATTGA CATGCTAGTT TTACGATTAC CGTTC ATCG ATTCTCTTGT TTGCTCCAGA CTCTCAGGCA ATGACCTGAT AGCCTTTGTA GATCTCTCAA AAATAGCTAC CCTCTC CGG CATTAATTTA TCAGCTAGAA CGGTTGAATA TCATATTGAT GGTGATTTGA CTGTCTCCGG CCTTTCTCAC CCTTTTG AA TCTTTACCTA CACATTACTC AGGCATTGCA TTTAAAATAT ATGAGGGTTC TAAAAATTTT TATCCTTGCG TTGAAATA A AGGCTTCTCC CGCAAAAGTA TTACAGGGTC ATAATGTTTT TGGTACAACC GATTTAGCTT TATGCTCTGA GGCTTTATT GCTTAATTTT GCTAATTCTT TGCCTTGCCT GTATGATTTA TTGGATGTT |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.04 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Acetate | |||||||||||||||
| Grid | Model: EMS / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number grids imaged: 1 / Number real images: 500 / Average exposure time: 1.0 sec. / Average electron dose: 11.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)