[English] 日本語
 Yorodumi
Yorodumi- EMDB-40642: Cryo-EM structure of the respiratory syncytial virus polymerase (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Respiratory syncytial virus / RNA-dependent RNA polymerase / VIRAL PROTEIN-RNA complex | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / Respiratory syncytial virus genome transcription / Translation of respiratory syncytial virus mRNAs / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Respiratory syncytial virus genome replication / RSV-host interactions / Assembly and release of respiratory syncytial virus (RSV) virions / Maturation of hRSV A proteins / Respiratory syncytial virus (RSV) attachment and entry ...NNS virus cap methyltransferase / Respiratory syncytial virus genome transcription / Translation of respiratory syncytial virus mRNAs / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Respiratory syncytial virus genome replication / RSV-host interactions / Assembly and release of respiratory syncytial virus (RSV) virions / Maturation of hRSV A proteins / Respiratory syncytial virus (RSV) attachment and entry / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Respiratory syncytial virus A2 Respiratory syncytial virus A2 | ||||||||||||||||||
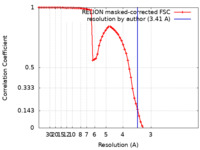
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | ||||||||||||||||||
 Authors Authors | Cao D / Gao Y / Chen Z / Gooneratne I / Roesler C / Mera C / Liang B | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structures of the promoter-bound respiratory syncytial virus polymerase. Authors: Dongdong Cao / Yunrong Gao / Zhenhang Chen / Inesh Gooneratne / Claire Roesler / Cristopher Mera / Paul D'Cunha / Anna Antonova / Deepak Katta / Sarah Romanelli / Qi Wang / Samantha Rice / ...Authors: Dongdong Cao / Yunrong Gao / Zhenhang Chen / Inesh Gooneratne / Claire Roesler / Cristopher Mera / Paul D'Cunha / Anna Antonova / Deepak Katta / Sarah Romanelli / Qi Wang / Samantha Rice / Wesley Lemons / Anita Ramanathan / Bo Liang /  Abstract: The respiratory syncytial virus (RSV) polymerase is a multifunctional RNA-dependent RNA polymerase composed of the large (L) protein and the phosphoprotein (P). It transcribes the RNA genome into ten ...The respiratory syncytial virus (RSV) polymerase is a multifunctional RNA-dependent RNA polymerase composed of the large (L) protein and the phosphoprotein (P). It transcribes the RNA genome into ten viral mRNAs and replicates full-length viral genomic and antigenomic RNAs. The RSV polymerase initiates RNA synthesis by binding to the conserved 3'-terminal RNA promoters of the genome or antigenome. However, the lack of a structure of the RSV polymerase bound to the RNA promoter has impeded the mechanistic understanding of RSV RNA synthesis. Here we report cryogenic electron microscopy structures of the RSV polymerase bound to its genomic and antigenomic viral RNA promoters, representing two of the first structures of an RNA-dependent RNA polymerase in complex with its RNA promoters in non-segmented negative-sense RNA viruses. The overall structures of the promoter-bound RSV polymerases are similar to that of the unbound (apo) polymerase. Our structures illustrate the interactions between the RSV polymerase and the RNA promoters and provide the structural basis for the initiation of RNA synthesis at positions 1 and 3 of the RSV promoters. These structures offer a deeper understanding of the pre-initiation state of the RSV polymerase and could aid in antiviral research against RSV. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40642.map.gz emd_40642.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40642-v30.xml emd-40642-v30.xml emd-40642.xml emd-40642.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40642_fsc.xml emd_40642_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40642.png emd_40642.png | 118.8 KB | ||
| Filedesc metadata |  emd-40642.cif.gz emd-40642.cif.gz | 8 KB | ||
| Others |  emd_40642_half_map_1.map.gz emd_40642_half_map_1.map.gz emd_40642_half_map_2.map.gz emd_40642_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40642 http://ftp.pdbj.org/pub/emdb/structures/EMD-40642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40642 | HTTPS FTP |
-Related structure data
| Related structure data |  8snyMC  8snxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40642.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40642.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40642_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40642_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The respiratory syncytial virus polymerase (L:P) bound to the tra...
| Entire | Name: The respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter |
|---|---|
| Components |
|
-Supramolecule #1: The respiratory syncytial virus polymerase (L:P) bound to the tra...
| Supramolecule | Name: The respiratory syncytial virus polymerase (L:P) bound to the trailer complementary promoter type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 250.704484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDPIINGNSA NVYLTDSYLK GVISFSECNA LGSYIFNGPY LKNDYTNLIS RQNPLIEHMN LKKLNITQSL ISKYHKGEIK LEEPTYFQS LLMTYKSMTS SEQIATTNLL KKIIRRAIEI SDVKVYAILN KLGLKEKDKI KSNNGQDEDN SVITTIIKDD I LSAVKDNQ ...String: MDPIINGNSA NVYLTDSYLK GVISFSECNA LGSYIFNGPY LKNDYTNLIS RQNPLIEHMN LKKLNITQSL ISKYHKGEIK LEEPTYFQS LLMTYKSMTS SEQIATTNLL KKIIRRAIEI SDVKVYAILN KLGLKEKDKI KSNNGQDEDN SVITTIIKDD I LSAVKDNQ SHLKADKNHS TKQKDTIKTT LLKKLMCSMQ HPPSWLIHWF NLYTKLNNIL TQYRSNEVKN HGFTLIDNQT LS GFQFILN QYGCIVYHKE LKRITVTTYN QFLTWKDISL SRLNVCLITW ISNCLNTLNK SLGLRCGFNN VILTQLFLYG DCI LKLFHN EGFYIIKEVE GFIMSLILNI TEEDQFRKRF YNSMLNNITD AANKAQKNLL SRVCHTLLDK TVSDNIINGR WIIL LSKFL KLIKLAGDNN LNNLSELYFL FRIFGHPMVD ERQAMDAVKI NCNETKFYLL SSLSMLRGAF IYRIIKGFVN NYNRW PTLR NAIVLPLRWL TYYKLNTYPS LLELTERDLI VLSGLRFYRE FRLPKKVDLE MIINDKAISP PKNLIWTSFP RNYMPS HIQ NYIEHEKLKF SESDKSRRVL EYYLRDNKFN ECDLYNCVVN QSYLNNPNHV VSLTGKEREL SVGRMFAMQP GMFRQVQ IL AEKMIAENIL QFFPESLTRY GDLELQKILE LKAGISNKSN RYNDNYNNYI SKCSIITDLS KFNQAFRYET SCICSDVL D ELHGVQSLFS WLHLTIPHVT IICTYRHAPP YIGDHIVDLN NVDEQSGLYR YHMGGIEGWC QKLWTIEAIS LLDLISLKG KFSITALING DNQSIDISKP IRLMEGQTHA QADYLLALNS LKLLYKEYAG IGHKLKGTET YISRDMQFMS KTIQHNGVYY PASIKKVLR VGPWINTILD DFKVSLESIG SLTQELEYRG ESLLCSLIFR NVWLYNQIAL QLKNHALCNN KLYLDILKVL K HLKTFFNL DNIDTALTLY MNLPMLFGGG DPNLLYRSFY RRTPDFLTEA IVHSVFILSY YTNHDLKDKL QDLSDDRLNK FL TCIITFD KNPNAEFVTL MRDPQALGSE RQAKITSEIN RLAVTEVLST APNKIFSKSA QHYTTTEIDL NDIMQNIEPT YPH GLRVVY ESLPFYKAEK IVNLISGTKS ITNILEKTSA IDLTDIDRAT EMMRKNITLL IRILPLDCNR DKREILSMEN LSIT ELSKY VRERSWSLSN IVGVTSPSIM YTMDIKYTTS TISSGIIIEK YNVNSLTRGE RGPTKPWVGS STQEKKTMPV YNRQV LTKK QRDQIDLLAK LDWVYASIDN KDEFMEELSI GTLGLTYEKA KKLFPQYLSV NYLHRLTVSS RPCEFPASIP AYRTTN YHF DTSPINRILT EKYGDEDIDI VFQNCISFGL SLMSVVEQFT NVCPNRIILI PKLNEIHLMK PPIFTGDVDI HKLKQVI QK QHMFLPDKIS LTQYVELFLS NKTLKSGSHV NSNLILAHKI SDYFHNTYIL STNLAGHWIL IIQLMKDSKG IFEKDWGE G YITDHMFINL KVFFNAYKTY LLCFHKGYGK AKLECDMNTS DLLCVLELID SSYWKSMSKV FLEQKVIKYI LSQDASLHR VKGCHSFKLW FLKRLNVAEF TVCPWVVNID YHPTHMKAIL TYIDLVRMGL INIDRIHIKN KHKFNDEFYT SNLFYINYNF SDNTHLLTK HIRIANSELE NNYNKLYHPT PETLENILAN PIKSNDKKTL NDYCIGKNVD SIMLPLLSNK KLIKSSAMIR T NYSKQDLY NLFPMVVIDR IIDHSGNTAK SNQLYTTTSH QISLVHNSTS LYCMLPWHHI NRFNFVFSST GCKISIEYIL KD LKIKDPN CIAFIGEGAG NLLLRTVVEL HPDIRYIYRS LKDCNDHSLP IEFLRLYNGH INIDYGENLT IPATDATNNI HWS YLHIKF AEPISLFVCD AELSVTVNWS KIIIEWSKHV RKCKYCSSVN KCMLIVKYHA QDDIDFKLDN ITILKTYVCL GSKL KGSEV YLVLTIGPAN IFPVFNVVQN AKLILSRTKN FIMPKKADKE SIDANIKSLI PFLCYPITKK GINTALSKLK SVVSG DILS YSIAGRNEVF SNKLINHKHM NILKWFNHVL NFRSTELNYN HLYMVESTYP YLSELLNSLT TNELKKLIKI TGSLLY NFH NE UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 27.165838 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD ...String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD AMVGLREEMI EKIRTEALMT NDRLEAMARL RNEESEKMAK DTSDEVSLNP TSEKLNNLLE GNDSDNDLSL ED F UniProtKB: Phosphoprotein |
-Macromolecule #3: RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3')
| Macromolecule | Name: RNA (5'-R(*UP*UP*UP*UP*UP*CP*UP*CP*GP*U)-3') / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 3.053772 KDa |
| Sequence | String: UUUUUCUCGU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.35 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.86 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)