+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | hPAD4 bound to inhibitory Fab hI365 | |||||||||||||||||||||
 Map data Map data | Relion sharpened map of PAD4 bound to inhibitory fab 365 | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | complex / deiminase / enzyme / arginine / inflammation / calcium binding / Fab / IMMUNE SYSTEM / HYDROLASE-IMMUNE SYSTEM complex | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhistone H3R2 arginine deiminase activity / histone H3R8 arginine deiminase activity / histone H3R17 arginine deiminase activity / histone arginine deiminase activity / histone H3R26 arginine deiminase activity / protein-arginine deiminase / protein-arginine deiminase activity / stem cell population maintenance / Chromatin modifying enzymes / post-translational protein modification ...histone H3R2 arginine deiminase activity / histone H3R8 arginine deiminase activity / histone H3R17 arginine deiminase activity / histone arginine deiminase activity / histone H3R26 arginine deiminase activity / protein-arginine deiminase / protein-arginine deiminase activity / stem cell population maintenance / Chromatin modifying enzymes / post-translational protein modification / protein modification process / nucleosome assembly / chromatin organization / chromatin remodeling / innate immune response / calcium ion binding / protein-containing complex / nucleoplasm / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
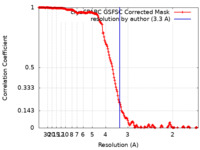
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||
 Authors Authors | Maker A / Verba KA | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4. Authors: Xin Zhou / Sophie Kong / Allison Maker / Soumya G Remesh / Kevin K Leung / Kliment A Verba / James A Wells /  Abstract: Unlocking the potential of protein arginine deiminase 4 (PAD4) as a drug target for rheumatoid arthritis requires a deeper understanding of its regulation. In this study, we use unbiased antibody ...Unlocking the potential of protein arginine deiminase 4 (PAD4) as a drug target for rheumatoid arthritis requires a deeper understanding of its regulation. In this study, we use unbiased antibody selections to identify functional antibodies capable of either activating or inhibiting PAD4 activity. Through cryogenic-electron microscopy, we characterized the structures of these antibodies in complex with PAD4 and revealed insights into their mechanisms of action. Rather than steric occlusion of the substrate-binding catalytic pocket, the antibodies modulate PAD4 activity through interactions with allosteric binding sites adjacent to the catalytic pocket. These binding events lead to either alteration of the active site conformation or the enzyme oligomeric state, resulting in modulation of PAD4 activity. Our study uses antibody engineering to reveal new mechanisms for enzyme regulation and highlights the potential of using PAD4 agonist and antagonist antibodies for studying PAD4-dependency in disease models and future therapeutic development. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40590.map.gz emd_40590.map.gz | 24.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40590-v30.xml emd-40590-v30.xml emd-40590.xml emd-40590.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40590_fsc.xml emd_40590_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_40590.png emd_40590.png | 112.8 KB | ||
| Masks |  emd_40590_msk_1.map emd_40590_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40590.cif.gz emd-40590.cif.gz | 6.5 KB | ||
| Others |  emd_40590_half_map_1.map.gz emd_40590_half_map_1.map.gz emd_40590_half_map_2.map.gz emd_40590_half_map_2.map.gz | 96.5 MB 96.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40590 http://ftp.pdbj.org/pub/emdb/structures/EMD-40590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40590 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40590 | HTTPS FTP |
-Validation report
| Summary document |  emd_40590_validation.pdf.gz emd_40590_validation.pdf.gz | 627.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40590_full_validation.pdf.gz emd_40590_full_validation.pdf.gz | 626.8 KB | Display | |
| Data in XML |  emd_40590_validation.xml.gz emd_40590_validation.xml.gz | 26.7 KB | Display | |
| Data in CIF |  emd_40590_validation.cif.gz emd_40590_validation.cif.gz | 35.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40590 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40590 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40590 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40590 | HTTPS FTP |
-Related structure data
| Related structure data |  8smlMC  8smkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40590.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40590.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion sharpened map of PAD4 bound to inhibitory fab 365 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40590_msk_1.map emd_40590_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of PAD4 bound to inhibitory fab 365
| File | emd_40590_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of PAD4 bound to inhibitory fab 365 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of PAD4 bound to inhibitory fab 365
| File | emd_40590_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of PAD4 bound to inhibitory fab 365 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : hPAD4 bound to inhibitory Fab hI365
| Entire | Name: hPAD4 bound to inhibitory Fab hI365 |
|---|---|
| Components |
|
-Supramolecule #1: hPAD4 bound to inhibitory Fab hI365
| Supramolecule | Name: hPAD4 bound to inhibitory Fab hI365 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 248 KDa |
-Macromolecule #1: Protein-arginine deiminase type-4
| Macromolecule | Name: Protein-arginine deiminase type-4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: protein-arginine deiminase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.902953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHASGG LNDIFEAQKI EWHEENLYFQ GTSAQGTLIR VTPEQPTHAV CVLGTLTQLD ICSSAPEDCT SFSINASPGV VVDIAHGPP AKKKSTGSST WPLDPGVEVT LTMKAASGST GDQKVQISYY GPKTPPVKAL LYLTGVEISL CADITRTGKV K PTRAVKDQ ...String: HHHHHHASGG LNDIFEAQKI EWHEENLYFQ GTSAQGTLIR VTPEQPTHAV CVLGTLTQLD ICSSAPEDCT SFSINASPGV VVDIAHGPP AKKKSTGSST WPLDPGVEVT LTMKAASGST GDQKVQISYY GPKTPPVKAL LYLTGVEISL CADITRTGKV K PTRAVKDQ RTWTWGPCGQ GAILLVNCDR DNLESSAMDC EDDEVLDSED LQDMSLMTLS TKTPKDFFTN HTLVLHVARS EM DKVRVFQ ATRGKLSSKC SVVLGPKWPS HYLMVPGGKH NMDFYVEALA FPDTDFPGLI TLTISLLDTS NLELPEAVVF QDS VVFRVA PWIMTPNTQP PQEVYACSIF ENEDFLKSVT TLAMKAKCKL TICPEEENMD DQWMQDEMEI GYIQAPHKTL PVVF DSPRN RGLKEFPIKR VMGPDFGYVT RGPQTGGISG LDSFGNLEVS PPVTVRGKEY PLGRILFGDS CYPSNDSRQM HQALQ DFLS AQQVQAPVKL YSDWLSVGHV DEFLSFVPAP DRKGFRLLLA SPRSCYKLFQ EQQNEGHGEA LLFEGIKKKK QQKIKN ILS NKTLREHNSF VERCIDWNRE LLKRELGLAE SDIIDIPQLF KLKEFSKAEA FFPNMVNMLV LGKHLGIPKP FGPVING RC CLEEKVCSLL EPLGLQCTFI NDFFTYHIRH GEVHCGTNVR RKPFSFKWWN MVP UniProtKB: Protein-arginine deiminase type-4 |
-Macromolecule #2: Fab hI365 light chain
| Macromolecule | Name: Fab hI365 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.157678 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QSSSSLVTFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSVS SAVAWYQQKP GKAPKLLIYS ASSLYSGVPS RFSGSRSGTD FTLTISSLQP EDFATYYCQ QSSSSLVTFG QGTKVEIKRT VAAPSVFIFP PSDSQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Fab hI365 heavy chain
| Macromolecule | Name: Fab hI365 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.615482 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFNFY YSIHWVRQAP GKGLEWVASI SPYSGYTSYA DSVKGRFTIS ADTSKNTAYL QMNSLRAED TAVYYCARKH PGSYPFWGWA LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EVQLVESGGG LVQPGGSLRL SCAASGFNFY YSIHWVRQAP GKGLEWVASI SPYSGYTSYA DSVKGRFTIS ADTSKNTAYL QMNSLRAED TAVYYCARKH PGSYPFWGWA LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK THT |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8sml: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)