+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human liver mitochondrial Superoxide dismutase [Mn] | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human / liver / mitochondrial / Superoxide dismutase [Mn] / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationacetylcholine-mediated vasodilation involved in regulation of systemic arterial blood pressure / erythrophore differentiation / : / negative regulation of membrane hyperpolarization / positive regulation of hydrogen peroxide biosynthetic process / response to magnetism / detection of oxygen / response to silicon dioxide / intracellular oxygen homeostasis / response to L-ascorbic acid ...acetylcholine-mediated vasodilation involved in regulation of systemic arterial blood pressure / erythrophore differentiation / : / negative regulation of membrane hyperpolarization / positive regulation of hydrogen peroxide biosynthetic process / response to magnetism / detection of oxygen / response to silicon dioxide / intracellular oxygen homeostasis / response to L-ascorbic acid / response to selenium ion / response to superoxide / response to manganese ion / hydrogen peroxide biosynthetic process / superoxide anion generation / intrinsic apoptotic signaling pathway in response to oxidative stress / positive regulation of vascular associated smooth muscle cell apoptotic process / response to zinc ion / superoxide metabolic process / superoxide dismutase / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / response to isolation stress / Detoxification of Reactive Oxygen Species / negative regulation of fat cell differentiation / superoxide dismutase activity / response to immobilization stress / hemopoiesis / negative regulation of vascular associated smooth muscle cell proliferation / mitochondrial nucleoid / response to electrical stimulus / cellular response to ethanol / response to hyperoxia / Mitochondrial unfolded protein response (UPRmt) / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / response to cadmium ion / neuron development / response to axon injury / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / negative regulation of fibroblast proliferation / glutathione metabolic process / removal of superoxide radicals / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / release of cytochrome c from mitochondria / response to activity / response to gamma radiation / regulation of mitochondrial membrane potential / post-embryonic development / respiratory electron transport chain / response to hydrogen peroxide / locomotory behavior / liver development / Transcriptional activation of mitochondrial biogenesis / oxygen binding / multicellular organismal-level iron ion homeostasis / regulation of blood pressure / intrinsic apoptotic signaling pathway in response to DNA damage / positive regulation of nitric oxide biosynthetic process / manganese ion binding / heart development / cellular response to oxidative stress / response to lipopolysaccharide / protein homotetramerization / negative regulation of neuron apoptotic process / response to hypoxia / positive regulation of cell migration / mitochondrial matrix / response to xenobiotic stimulus / negative regulation of cell population proliferation / regulation of transcription by RNA polymerase II / enzyme binding / mitochondrion / DNA binding / extracellular exosome / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
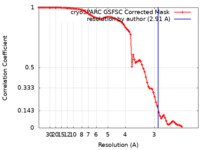
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Zhang Z / Tringides M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell Proteomics / Year: 2023 Journal: Mol Cell Proteomics / Year: 2023Title: High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology. Authors: Zhemin Zhang / Marios L Tringides / Christopher E Morgan / Masaru Miyagi / Jason A Mears / Charles L Hoppel / Edward W Yu /  Abstract: The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of ...The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of single particle cryo-EM to identify multiple proteins from three enriched heterogeneous fractions prepared from human liver mitochondrial lysate. We simultaneously identify and solve high-resolution structures of nine essential mitochondrial enzymes with key metabolic functions, including fatty acid catabolism, reactive oxidative species clearance, and amino acid metabolism. Our methodology also identified multiple distinct members of the acyl-CoA dehydrogenase family. This work highlights the potential of cryo-EM to explore tissue proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40566.map.gz emd_40566.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40566-v30.xml emd-40566-v30.xml emd-40566.xml emd-40566.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40566_fsc.xml emd_40566_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_40566.png emd_40566.png | 144.3 KB | ||
| Filedesc metadata |  emd-40566.cif.gz emd-40566.cif.gz | 5.4 KB | ||
| Others |  emd_40566_half_map_1.map.gz emd_40566_half_map_1.map.gz emd_40566_half_map_2.map.gz emd_40566_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40566 http://ftp.pdbj.org/pub/emdb/structures/EMD-40566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40566 | HTTPS FTP |
-Related structure data
| Related structure data |  8sksMC  8sgpC  8sgrC  8sgsC  8sgvC  8shsC  8sk6C  8sk8C  8skrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40566.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40566.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Superoxide dismutase [Mn]
| Entire | Name: Superoxide dismutase [Mn] |
|---|---|
| Components |
|
-Supramolecule #1: Superoxide dismutase [Mn]
| Supramolecule | Name: Superoxide dismutase [Mn] / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Superoxide dismutase [Mn], mitochondrial
| Macromolecule | Name: Superoxide dismutase [Mn], mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: superoxide dismutase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.782119 KDa |
| Sequence | String: MLSRAVCGTS RQLAPVLGYL GSRQKHSLPD LPYDYGALEP HINAQIMQLH HSKHHAAYVN NLNVTEEKYQ EALAKGDVTA QIALQPALK FNGGGHINHS IFWTNLSPNG GGEPKGELLE AIKRDFGSFD KFKEKLTAAS VGVQGSGWGW LGFNKERGHL Q IAACPNQD ...String: MLSRAVCGTS RQLAPVLGYL GSRQKHSLPD LPYDYGALEP HINAQIMQLH HSKHHAAYVN NLNVTEEKYQ EALAKGDVTA QIALQPALK FNGGGHINHS IFWTNLSPNG GGEPKGELLE AIKRDFGSFD KFKEKLTAAS VGVQGSGWGW LGFNKERGHL Q IAACPNQD PLQGTTGLIP LLGIDVWEHA YYLQYKNVRP DYLKAIWNVI NWENVTERYM ACKK UniProtKB: Superoxide dismutase [Mn], mitochondrial |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 39.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)